RNA degradation in Saccharomyces cerevisae
- PMID: 22785621
- PMCID: PMC3389967
- DOI: 10.1534/genetics.111.137265
RNA degradation in Saccharomyces cerevisae
Abstract
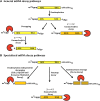
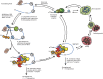
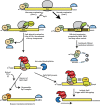
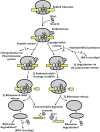
All RNA species in yeast cells are subject to turnover. Work over the past 20 years has defined degradation mechanisms for messenger RNAs, transfer RNAs, ribosomal RNAs, and noncoding RNAs. In addition, numerous quality control mechanisms that target aberrant RNAs have been identified. Generally, each decay mechanism contains factors that funnel RNA substrates to abundant exo- and/or endonucleases. Key issues for future work include determining the mechanisms that control the specificity of RNA degradation and how RNA degradation processes interact with translation, RNA transport, and other cellular processes.
Figures
Similar articles
-
Sequential RNA degradation pathways provide a fail-safe mechanism to limit the accumulation of unspliced transcripts in Saccharomyces cerevisiae.RNA. 2012 Aug;18(8):1563-72. doi: 10.1261/rna.033779.112. Epub 2012 Jul 2. RNA. 2012. PMID: 22753783 Free PMC article.
-
Nonsense-containing mRNAs that accumulate in the absence of a functional nonsense-mediated mRNA decay pathway are destabilized rapidly upon its restitution.Mol Cell Biol. 2003 Feb;23(3):842-51. doi: 10.1128/MCB.23.3.842-851.2003. Mol Cell Biol. 2003. PMID: 12529390 Free PMC article.
-
In Vivo Mapping of Eukaryotic RNA Interactomes Reveals Principles of Higher-Order Organization and Regulation.Mol Cell. 2016 May 19;62(4):603-17. doi: 10.1016/j.molcel.2016.04.028. Epub 2016 May 12. Mol Cell. 2016. PMID: 27184079
-
Molecular chaperones and quality control in noncoding RNA biogenesis.Cold Spring Harb Symp Quant Biol. 2006;71:505-11. doi: 10.1101/sqb.2006.71.051. Cold Spring Harb Symp Quant Biol. 2006. PMID: 17381333 Review.
-
Mechanisms and control of mRNA decapping in Saccharomyces cerevisiae.Annu Rev Biochem. 2000;69:571-95. doi: 10.1146/annurev.biochem.69.1.571. Annu Rev Biochem. 2000. PMID: 10966469 Review.
Cited by
-
PELOTA and HBS1 suppress co-translational messenger RNA decay in Arabidopsis.Plant Direct. 2023 Dec 26;7(12):e553. doi: 10.1002/pld3.553. eCollection 2023 Dec. Plant Direct. 2023. PMID: 38149303 Free PMC article.
-
Splicing-Mediated Autoregulation Modulates Rpl22p Expression in Saccharomyces cerevisiae.PLoS Genet. 2016 Apr 20;12(4):e1005999. doi: 10.1371/journal.pgen.1005999. eCollection 2016 Apr. PLoS Genet. 2016. PMID: 27097027 Free PMC article.
-
A trans-dominant form of Gag restricts Ty1 retrotransposition and mediates copy number control.J Virol. 2015 Apr;89(7):3922-38. doi: 10.1128/JVI.03060-14. Epub 2015 Jan 21. J Virol. 2015. PMID: 25609815 Free PMC article.
-
Messenger RNAs of Yeast Virus-Like Elements Contain Non-templated 5' Poly(A) Leaders, and Their Expression Is Independent of eIF4E and Pab1.Front Microbiol. 2019 Oct 30;10:2366. doi: 10.3389/fmicb.2019.02366. eCollection 2019. Front Microbiol. 2019. PMID: 31736885 Free PMC article.
-
The Ess1 prolyl isomerase: traffic cop of the RNA polymerase II transcription cycle.Biochim Biophys Acta. 2014;1839(4):316-33. doi: 10.1016/j.bbagrm.2014.02.001. Epub 2014 Feb 12. Biochim Biophys Acta. 2014. PMID: 24530645 Free PMC article. Review.
References
-
- Aebi M., Kirchner G., Chen J. Y., Vijayraghavan U., Jacobson A., et al. , 1990. Isolation of a temperature-sensitive mutant with an altered tRNA nucleotidyltransferase and cloning of the gene encoding tRNA nucleotidyltransferase in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 265: 16216–16220 - PubMed
-
- Alexandrov A., Chernyakov I., Gu W., Hiley S. L., Hughes T. R., et al. , 2006. Rapid tRNA decay can result from lack of nonessential modifications. Mol. Cell 21: 87–96 - PubMed
-
- Amrani N., Ganesan R., Kervestin S., Mangus D. A., Ghosh S., et al. , 2004. A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature 432: 112–118 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous