SLC33A1/AT-1 protein regulates the induction of autophagy downstream of IRE1/XBP1 pathway
- PMID: 22787145
- PMCID: PMC3436137
- DOI: 10.1074/jbc.M112.363911
SLC33A1/AT-1 protein regulates the induction of autophagy downstream of IRE1/XBP1 pathway
Abstract
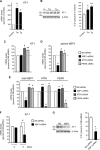
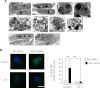
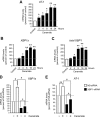
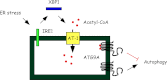
One of the main functions of the unfolded protein response is to ensure disposal of large protein aggregates that accumulate in the lumen of the endoplasmic reticulum (ER) whereas avoiding, at least under nonlethal levels of ER stress, cell death. When tightly controlled, autophagy-dependent ER-associated degradation (ERAD(II)) allows the cell to recover from the transient accumulation of protein aggregates; however, when unchecked, it can be detrimental and cause autophagic cell death/type 2 cell death. Here we show that IRE1/XBP1 controls the induction of autophagy/ERAD(II) during the unfolded protein response by activating the ER membrane transporter SLC33A1/AT-1, which ensures continuous supply of acetyl-CoA into the lumen of the ER. Failure to induce AT-1 leads to widespread autophagic cell death. Mechanistically, the regulation of the autophagic process involves N(ε)-lysine acetylation of Atg9A.
Figures


References
-
- Trombetta E. S., Parodi A. J. (2003) Quality control and protein folding in the secretory pathway. Annu. Rev. Cell Dev. Biol. 19, 649–676 - PubMed
-
- Kleizen B., Braakman I. (2004) Protein folding and quality control in the endoplasmic reticulum. Curr. Opin. Cell Biol. 16, 343–349 - PubMed
-
- Buchberger A., Bukau B., Sommer T. (2010) Protein quality control in the cytosol and the endoplasmic reticulum: brothers in arms. Mol. Cell 40, 238–252 - PubMed
-
- Klionsky D. J. (2006) Neurodegeneration: good riddance to bad rubbish. Nature 441, 819–820 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

