A member of a new Picornaviridae genus is shed in pig feces
- PMID: 22787214
- PMCID: PMC3446573
- DOI: 10.1128/JVI.00046-12
A member of a new Picornaviridae genus is shed in pig feces
Abstract
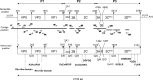
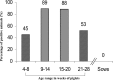
During a study of the fecal microbiomes from two healthy piglets using high-throughput sequencing (HTS), we identified a viral genome containing an open reading frame encoding a predicted polyprotein of 2,133 amino acids. This novel viral genome displayed the typical organization of picornaviruses, containing three structural proteins (VP0, VP3, and VP1), followed by seven nonstructural proteins (2A, 2B, 2C, 3A, 3B, 3C(pro), and 3D(pol)). Given its particular relationship with Parechovirus, we propose to name it "Pasivirus" for Parecho sister clade virus, with "Swine pasivirus 1" (SPaV1) as the type species. Fecal samples collected at an industrial farm from healthy sows and piglets from the same herd (25 and 75, respectively) with ages ranging from 4 to 28 weeks were analyzed for the presence of SPaV1 by one-step reverse transcription (RT)-PCR targeting a 3D region of 151 bp. SPaV1 was detected in fecal samples from 51/75 healthy piglets (68% of the animals) and in none of the 25 fecal samples from healthy sows, indicating that SPaV1 circulates through enteric infection of healthy piglets. We propose that SPaV1 represents the first member of a novel Picornaviridae genus related to parechoviruses.
Figures





References
-
- Ambros V, Baltimore D. 1978. Protein is linked to the 5′ end of poliovirus RNA by a phosphodiester linkage to tyrosine. J. Biol. Chem. 253:5263–5266 - PubMed
-
- Barbknecht M. 2009. Characterization of an unclassified virus and survey for its presence in Wisconsin bluegill populations. M.Sc. thesis University of Wisconsin—La Crosse, La Crosse, WI
-
- Beckett R, Miller WA. 2007. Rapid full-length cloning of nonpolyadenylated RNA virus genomes. Curr. Protoc. Microbiol. 16:F.3.1–F.3.18 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

