A multivalent adsorption apparatus explains the broad host range of phage phi92: a comprehensive genomic and structural analysis
- PMID: 22787233
- PMCID: PMC3457257
- DOI: 10.1128/JVI.00801-12
A multivalent adsorption apparatus explains the broad host range of phage phi92: a comprehensive genomic and structural analysis
Abstract
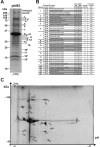
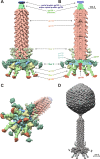
Bacteriophage phi92 is a large, lytic myovirus isolated in 1983 from pathogenic Escherichia coli strains that carry a polysialic acid capsule. Here we report the genome organization of phi92, the cryoelectron microscopy reconstruction of its virion, and the reinvestigation of its host specificity. The genome consists of a linear, double-stranded 148,612-bp DNA sequence containing 248 potential open reading frames and 11 putative tRNA genes. Orthologs were found for 130 of the predicted proteins. Most of the virion proteins showed significant sequence similarities to proteins of myoviruses rv5 and PVP-SE1, indicating that phi92 is a new member of the novel genus of rv5-like phages. Reinvestigation of phi92 host specificity showed that the host range is not limited to polysialic acid-encapsulated Escherichia coli but includes most laboratory strains of Escherichia coli and many Salmonella strains. Structure analysis of the phi92 virion demonstrated the presence of four different types of tail fibers and/or tailspikes, which enable the phage to use attachment sites on encapsulated and nonencapsulated bacteria. With this report, we provide the first detailed description of a multivalent, multispecies phage armed with a host cell adsorption apparatus resembling a nanosized Swiss army knife. The genome, structure, and, in particular, the organization of the baseplate of phi92 demonstrate how a bacteriophage can evolve into a multi-pathogen-killing agent.
Figures



References
-
- Baker TS, Henderson R. 2006. Electron cryomicroscopy, p 451–463 In International tables for crystallography, vol. F Springer, New York, NY
-
- Barry GT, Goebel WF. 1957. Colominic acid, a substance of bacterial origin related to sialic acid. Nature 179:206. - PubMed
-
- Bateman A, Eddy SR, Mesyanzhinov VV. 1997. A member of the immunoglobulin superfamily in bacteriophage T4. Virus Genes 14:163–165 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

