Inhibition of RNA polymerase I as a therapeutic strategy to promote cancer-specific activation of p53
- PMID: 22789538
- PMCID: PMC3749732
- DOI: 10.1016/j.ccr.2012.05.019
Inhibition of RNA polymerase I as a therapeutic strategy to promote cancer-specific activation of p53
Abstract
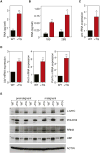
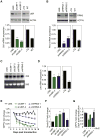
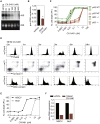
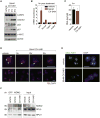
Increased transcription of ribosomal RNA genes (rDNA) by RNA Polymerase I is a common feature of human cancer, but whether it is required for the malignant phenotype remains unclear. We show that rDNA transcription can be therapeutically targeted with the small molecule CX-5461 to selectively kill B-lymphoma cells in vivo while maintaining a viable wild-type B cell population. The therapeutic effect is a consequence of nucleolar disruption and activation of p53-dependent apoptotic signaling. Human leukemia and lymphoma cell lines also show high sensitivity to inhibition of rDNA transcription that is dependent on p53 mutational status. These results identify selective inhibition of rDNA transcription as a therapeutic strategy for the cancer specific activation of p53 and treatment of hematologic malignancies.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures

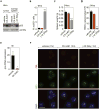

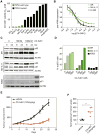
References
-
- Adams JM, Harris AW, Pinkert CA, Corcoran LM, Alexander WS, Cory S, Palmiter RD, Brinster RL. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985;318:533–538. - PubMed
-
- Arabi A, Wu S, Ridderstråle K, Bierhoff H, Shiue C, Fatyol K, Fahlén S, Hydbring P, Söderberg O, Grummt I, et al. c-Myc associates with ribosomal DNA and activates RNA polymerase I transcription. Nat Cell Biol. 2005;7:303–310. - PubMed
-
- Boisvert FM, van Koningsbruggen S, Navascués J, Lamond AI. The multifunctional nucleolus. Nat Rev Mol Cell Biol. 2007;8:574–585. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

