A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2
- PMID: 22789542
- PMCID: PMC3954744
- DOI: 10.1016/j.ccr.2012.05.015
A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2
Abstract
Genes mutated in patients with Fanconi anemia (FA) interact with the DNA repair genes BRCA1 and BRCA2/FANCD1 to suppress tumorigenesis, but the molecular functions ascribed to them cannot fully explain all of their cellular roles. Here, we show a repair-independent requirement for FA genes, including FANCD2, and BRCA1 in protecting stalled replication forks from degradation. Fork protection is surprisingly rescued in FANCD2-deficient cells by elevated RAD51 levels or stabilized RAD51 filaments. Moreover, FANCD2-mediated fork protection is epistatic with RAD51 functions, revealing an unanticipated fork protection pathway that connects FA genes to RAD51 and the BRCA1/2 breast cancer suppressors. Collective results imply a unified molecular mechanism for repair-independent functions of FA, RAD51, and BRCA1/2 proteins in preventing genomic instability and suppressing tumorigenesis.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures
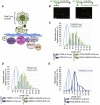
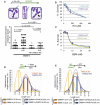
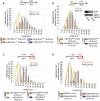
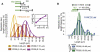


Comment in
-
To the rescue: the Fanconi anemia genome stability pathway salvages replication forks.Cancer Cell. 2012 Jul 10;22(1):5-6. doi: 10.1016/j.ccr.2012.06.006. Cancer Cell. 2012. PMID: 22789533 Free PMC article.
References
-
- Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N, Vassiliou LV, Kolettas E, Niforou K, Zoumpourlis VC, et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444:633–637. - PubMed
-
- Chen J, Silver DP, Walpita D, Cantor SB, Gazdar AF, Tomlinson G, Couch FJ, Weber BL, Ashley T, Livingston DM, Scully R. Stable interaction between the products of the BRCA1 and BRCA2 tumor suppressor genes in mitotic and meiotic cells. Mol. Cell. 1998;2:317–328. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

