Rotavirus non-structural proteins: structure and function
- PMID: 22789743
- PMCID: PMC3422752
- DOI: 10.1016/j.coviro.2012.06.003
Rotavirus non-structural proteins: structure and function
Abstract
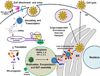
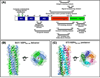
The replication of rotavirus is a complex process that is orchestrated by an exquisite interplay between the rotavirus non-structural and structural proteins. Subsequent to particle entry and genome transcription, the non-structural proteins coordinate and regulate viral mRNA translation and the formation of electron-dense viroplasms that serve as exclusive compartments for genome replication, genome encapsidation and capsid assembly. In addition, non-structural proteins are involved in antagonizing the antiviral host response and in subverting important cellular processes to enable successful virus replication. Although far from complete, new structural studies, together with functional studies, provide substantial insight into how the non-structural proteins coordinate rotavirus replication. This brief review highlights our current knowledge of the structure-function relationships of the rotavirus non-structural proteins, as well as fascinating questions that remain to be understood.
Copyright © 2012 Elsevier B.V. All rights reserved.
Figures

Similar articles
-
Recombinant Rotaviruses Rescued by Reverse Genetics Reveal the Role of NSP5 Hyperphosphorylation in the Assembly of Viral Factories.J Virol. 2019 Dec 12;94(1):e01110-19. doi: 10.1128/JVI.01110-19. Print 2019 Dec 12. J Virol. 2019. PMID: 31619556 Free PMC article.
-
Viroplasms: Assembly and Functions of Rotavirus Replication Factories.Viruses. 2021 Jul 12;13(7):1349. doi: 10.3390/v13071349. Viruses. 2021. PMID: 34372555 Free PMC article. Review.
-
The Role of the Host Cytoskeleton in the Formation and Dynamics of Rotavirus Viroplasms.Viruses. 2024 Apr 25;16(5):668. doi: 10.3390/v16050668. Viruses. 2024. PMID: 38793550 Free PMC article. Review.
-
Emerging themes in rotavirus cell entry, genome organization, transcription and replication.Virus Res. 2004 Apr;101(1):67-81. doi: 10.1016/j.virusres.2003.12.007. Virus Res. 2004. PMID: 15010218 Review.
-
COPII Vesicle Transport Is Required for Rotavirus NSP4 Interaction with the Autophagy Protein LC3 II and Trafficking to Viroplasms.J Virol. 2019 Dec 12;94(1):e01341-19. doi: 10.1128/JVI.01341-19. Print 2019 Dec 12. J Virol. 2019. PMID: 31597778 Free PMC article.
Cited by
-
Crystal structure and nucleic acid binding mode of CPV NSP9: implications for viroplasm in Reovirales.Nucleic Acids Res. 2024 Oct 14;52(18):11115-11127. doi: 10.1093/nar/gkae803. Nucleic Acids Res. 2024. PMID: 39287123 Free PMC article.
-
Identification of a Ruminant Origin Group B Rotavirus Associated with Diarrhea Outbreaks in Foals.Viruses. 2021 Jul 9;13(7):1330. doi: 10.3390/v13071330. Viruses. 2021. PMID: 34372536 Free PMC article.
-
Intercellular Transport of Viral Proteins.Results Probl Cell Differ. 2024;73:435-474. doi: 10.1007/978-3-031-62036-2_18. Results Probl Cell Differ. 2024. PMID: 39242389 Review.
-
Mutational analysis of the rotavirus NSP4 enterotoxic domain that binds to caveolin-1.Virol J. 2013 Nov 13;10:336. doi: 10.1186/1743-422X-10-336. Virol J. 2013. PMID: 24220211 Free PMC article.
-
Formulation and preclinical studies with a trivalent rotavirus P2-VP8 subunit vaccine.Hum Vaccin Immunother. 2020 Aug 2;16(8):1957-1968. doi: 10.1080/21645515.2019.1710412. Epub 2020 Jan 29. Hum Vaccin Immunother. 2020. PMID: 31995444 Free PMC article.
References
-
- Estes MK, Greenberg HB. Rotaviruses. In: Knipe PMH DM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE, editors. Fields Virology. edn 6th. Lippincott Williams & Wilkins; 2012. In press.
-
- Mattion NM, Mitchell DB, Both GW, Estes MK. Expression of rotavirus proteins encoded by alternative open reading frames of genome segment 11. Virology. 1991;181:295–304. - PubMed
-
- Lawton JA, Estes MK, Prasad BV. Three-dimensional visualization of mRNA release from actively transcribing rotavirus particles. Nat Struct Biol. 1997;4:118–121. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

