Insights into p53 transcriptional function via genome-wide chromatin occupancy and gene expression analysis
- PMID: 22790872
- PMCID: PMC3504713
- DOI: 10.1038/cdd.2012.89
Insights into p53 transcriptional function via genome-wide chromatin occupancy and gene expression analysis
Abstract
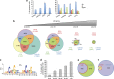
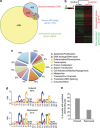
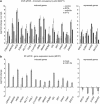
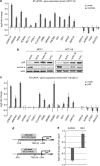
The tumor-suppressor p53 can induce various biological responses. Yet, it is not clear whether it is p53 in vivo promoter selectivity that triggers different transcription programs leading to different outcomes. Our analysis of genome-wide chromatin occupancy by p53 using chromatin immunoprecipitation (ChIP)-seq revealed 'p53 default program', that is, the pattern of major p53-bound sites that is similar upon p53 activation by nutlin3a, reactivation of p53 and induction of tumor cell apoptosis (RITA) or 5-fluorouracil in breast cancer cells, despite different biological outcomes. Parallel analysis of gene expression allowed identification of 280 novel p53 target genes, including p53-repressed AURKA. We identified Sp1 as one of the p53 modulators, which confer specificity to p53-mediated transcriptional response upon RITA. Further, we found that STAT3 antagonizes p53-mediated repression of a subset of genes, including AURKA.
Figures

References
-
- Goh AM, Coffill CR, Lane DP. The role of mutant p53 in human cancer. J Pathol. 2011;223:116–126. - PubMed
-
- Vousden KH, Prives C. Blinded by the light: the growing complexity of p53. Cell. 2009;137:413–431. - PubMed
-
- Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. - PubMed
-
- Issaeva N, Bozko P, Enge M, Protopopova M, Verhoef LG, Masucci M, et al. Small molecule RITA binds to p53, blocks p53-HDM-2 interaction and activates p53 function in tumors. Nat Med. 2004;10:1321–1328. - PubMed
-
- Ding K, Lu Y, Nikolovska-Coleska Z, Wang G, Qiu S, Shangary S, et al. Structure-based design of spiro-oxindoles as potent, specific small-molecule inhibitors of the MDM2-p53 interaction. J Med Chem. 2006;49:3432–3435. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

