Protein-SIP enables time-resolved analysis of the carbon flux in a sulfate-reducing, benzene-degrading microbial consortium
- PMID: 22791237
- PMCID: PMC3504967
- DOI: 10.1038/ismej.2012.68
Protein-SIP enables time-resolved analysis of the carbon flux in a sulfate-reducing, benzene-degrading microbial consortium
Abstract
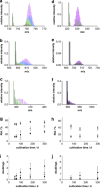
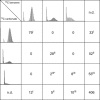
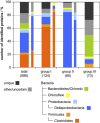
Benzene is a major contaminant in various environments, but the mechanisms behind its biodegradation under strictly anoxic conditions are not yet entirely clear. Here we analyzed a benzene-degrading, sulfate-reducing enrichment culture originating from a benzene-contaminated aquifer by a metagenome-based functional metaproteomic approach, using protein-based stable isotope probing (protein-SIP). The time-resolved, quantitative analysis of carbon fluxes within the community supplied with either (13)C-labeled benzene or (13)C-labeled carbonate yielded different functional groups of organisms, with their peptides showing specific time dependencies of (13)C relative isotope abundance indicating different carbon utilization. Through a detailed analysis of the mass spectrometric (MS) data, it was possible to quantify the utilization of the initial carbon source and the metabolic intermediates. The functional groups were affiliated to Clostridiales, Deltaproteobacteria and Bacteroidetes/Chlorobi. The Clostridiales-related organisms were involved in benzene degradation, putatively by fermentation, and additionally used significant amounts of carbonate as a carbon source. The other groups of organisms were found to perform diverse functions, with Deltaproteobacteria degrading fermentation products and Bacteroidetes/Chlorobi being putative scavengers feeding on dead cells. A functional classification of identified proteins supported this allocation and gave further insights into the metabolic pathways and the interactions between the community members. This example shows how protein-SIP can be applied to obtain temporal and phylogenetic information about functional interdependencies within microbial communities.
Figures


References
-
- Bastida F, Rosell M, Franchini AG, Seifert J, Finsterbusch S, Jehmlich N, et al. Elucidating MTBE degradation in a mixed consortium using a multidisciplinary approach. FEMS Microbiol Ecol. 2010;73:370–384. - PubMed
-
- Boschker HTS, Nold SC, Wellsbury P, Bos D, de Graaf W, Pel R, et al. Direct linking of microbial populations to specific biogeochemical processes by C-13-labelling of biomarkers. Nature. 1998;392:801–805.
-
- Chen Y, Murrell JC. When metagenomics meets stable-isotope probing: progress and perspectives. Trends Microbiol. 2010;18:157–163. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

