Toward stable genetic engineering of human O-glycosylation in plants
- PMID: 22791304
- PMCID: PMC3440218
- DOI: 10.1104/pp.112.198200
Toward stable genetic engineering of human O-glycosylation in plants
Abstract
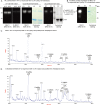
Glycosylation is the most abundant and complex posttranslational modification to be considered for recombinant production of therapeutic proteins. Mucin-type (N-acetylgalactosamine [GalNAc]-type) O-glycosylation is found in eumetazoan cells but absent in plants and yeast, making these cell types an obvious choice for de novo engineering of this O-glycosylation pathway. We previously showed that transient implementation of O-glycosylation capacity in plants requires introduction of the synthesis of the donor substrate UDP-GalNAc and one or more polypeptide GalNAc-transferases for incorporating GalNAc residues into proteins. Here, we have stably engineered O-glycosylation capacity in two plant cell systems, soil-grown Arabidopsis (Arabidopsis thaliana) and tobacco (Nicotiana tabacum) Bright Yellow-2 suspension culture cells. Efficient GalNAc O-glycosylation of two stably coexpressed substrate O-glycoproteins was obtained, but a high degree of proline hydroxylation and hydroxyproline-linked arabinosides, on a mucin (MUC1)-derived substrate, was also observed. Addition of the prolyl 4-hydroxylase inhibitor 2,2-dipyridyl, however, effectively suppressed proline hydroxylation and arabinosylation of MUC1 in Bright Yellow-2 cells. In summary, stably engineered mammalian type O-glycosylation was established in transgenic plants, demonstrating that plants may serve as host cells for the production of recombinant O-glycoproteins. However, the present stable implementation further strengthens the notion that elimination of endogenous posttranslational modifications may be needed for the production of protein therapeutics.
Figures





References
-
- Altmann F. (2007) The role of protein glycosylation in allergy. Int Arch Allergy Immunol 142: 99–115 - PubMed
-
- Bennett EP, Hassan H, Mandel U, Mirgorodskaya E, Roepstorff P, Burchell J, Taylor-Papadimitriou J, Hollingsworth MA, Merkx G, van Kessel AG, et al. (1998) Cloning of a human UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase that complements other GalNAc-transferases in complete O-glycosylation of the MUC1 tandem repeat. J Biol Chem 273: 30472–30481 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

