Proteobacterial ArfA peptides are synthesized from non-stop messenger RNAs
- PMID: 22791716
- PMCID: PMC3436202
- DOI: 10.1074/jbc.M112.374074
Proteobacterial ArfA peptides are synthesized from non-stop messenger RNAs
Abstract
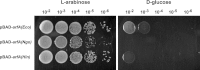
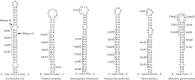
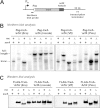
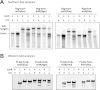
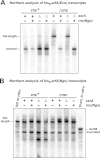
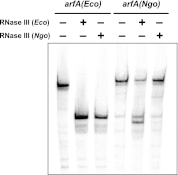
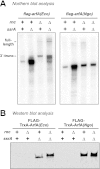
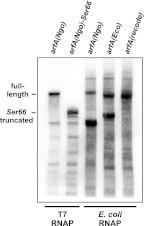
The translation of non-stop mRNA (which lack in-frame stop codons) represents a significant quality control problem for all organisms. In eubacteria, the transfer-messenger RNA (tmRNA) system facilitates recycling of stalled ribosomes from non-stop mRNA in a process termed trans-translation or ribosome rescue. During rescue, the nascent chain is tagged with the tmRNA-encoded ssrA peptide, which promotes polypeptide degradation after release from the stalled ribosome. Escherichia coli possesses an additional ribosome rescue pathway mediated by the ArfA peptide. The E. coli arfA message contains a hairpin structure that is cleaved by RNase III to produce a non-stop transcript. Therefore, ArfA levels are controlled by tmRNA through ssrA-peptide tagging and proteolysis. Here, we examine whether ArfA homologues from other bacteria are also regulated by RNase III and tmRNA. We searched 431 arfA coding sequences for mRNA secondary structures and found that 82.8% of the transcripts contain predicted hairpins in their 3'-coding regions. The arfA hairpins from Haemophilus influenzae, Proteus mirabilis, Vibrio fischeri, and Pasteurella multocida are all cleaved by RNase III as predicted, whereas the hairpin from Neisseria gonorrhoeae functions as an intrinsic transcription terminator to generate non-stop mRNA. Each ArfA homologue is ssrA-tagged and degraded when expressed in wild-type E. coli cells, but accumulates in mutants lacking tmRNA. Together, these findings show that ArfA synthesis from non-stop mRNA is a conserved mechanism to regulate the alternative ribosome rescue pathway. This strategy ensures that ArfA homologues are only deployed when the tmRNA system is incapacitated or overwhelmed by stalled ribosomes.
Figures
References
-
- Moore S. D., Sauer R. T. (2007) The tmRNA system for translational surveillance and ribosome rescue. Annu. Rev. Biochem. 76, 101–124 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

