Prediction of microbial growth rate versus biomass yield by a metabolic network with kinetic parameters
- PMID: 22792053
- PMCID: PMC3390398
- DOI: 10.1371/journal.pcbi.1002575
Prediction of microbial growth rate versus biomass yield by a metabolic network with kinetic parameters
Abstract
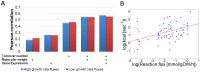
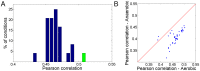
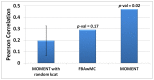
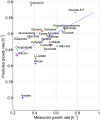
Identifying the factors that determine microbial growth rate under various environmental and genetic conditions is a major challenge of systems biology. While current genome-scale metabolic modeling approaches enable us to successfully predict a variety of metabolic phenotypes, including maximal biomass yield, the prediction of actual growth rate is a long standing goal. This gap stems from strictly relying on data regarding reaction stoichiometry and directionality, without accounting for enzyme kinetic considerations. Here we present a novel metabolic network-based approach, MetabOlic Modeling with ENzyme kineTics (MOMENT), which predicts metabolic flux rate and growth rate by utilizing prior data on enzyme turnover rates and enzyme molecular weights, without requiring measurements of nutrient uptake rates. The method is based on an identified design principle of metabolism in which enzymes catalyzing high flux reactions across different media tend to be more efficient in terms of having higher turnover numbers. Extending upon previous attempts to utilize kinetic data in genome-scale metabolic modeling, our approach takes into account the requirement for specific enzyme concentrations for catalyzing predicted metabolic flux rates, considering isozymes, protein complexes, and multi-functional enzymes. MOMENT is shown to significantly improve the prediction accuracy of various metabolic phenotypes in E. coli, including intracellular flux rates and changes in gene expression levels under different growth rates. Most importantly, MOMENT is shown to predict growth rates of E. coli under a diverse set of media that are correlated with experimental measurements, markedly improving upon existing state-of-the art stoichiometric modeling approaches. These results support the view that a physiological bound on cellular enzyme concentrations is a key factor that determines microbial growth rate.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
-
- Lengauer T, editor. Bioinformatics - From Genomes to Therapies 2. Getting at the Inner Workings: Molecular Interactions. Weinheim, Germany: Wiley-VCH; 2007. 520
-
- Price ND, Papin JA, Schilling CH, Palsson BO. Genome-scale microbial in silico models: the constraints-based approach. Trends Biotechnol. 2003;21:162–169. - PubMed
-
- Stelling J, Klamt S, Bettenbrock K, Schuster S, Gilles ED. Metabolic network structure determines key aspects of functionality and regulation. Nature. 2002;420:190–193. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

