Predicting signatures of "synthetic associations" and "natural associations" from empirical patterns of human genetic variation
- PMID: 22792059
- PMCID: PMC3390358
- DOI: 10.1371/journal.pcbi.1002600
Predicting signatures of "synthetic associations" and "natural associations" from empirical patterns of human genetic variation
Abstract
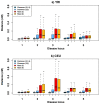
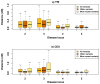
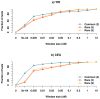
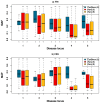
Genome-wide association studies (GWAS) have in recent years discovered thousands of associated markers for hundreds of phenotypes. However, associated loci often only explain a relatively small fraction of heritability and the link between association and causality has yet to be uncovered for most loci. Rare causal variants have been suggested as one scenario that may partially explain these shortcomings. Specifically, Dickson et al. recently reported simulations of rare causal variants that lead to association signals of common, tag single nucleotide polymorphisms, dubbed "synthetic associations". However, an open question is what practical implications synthetic associations have for GWAS. Here, we explore the signatures exhibited by such "synthetic associations" and their implications based on patterns of genetic variation observed in human populations, thus accounting for human evolutionary history -a force disregarded in previous simulation studies. This is made possible by human population genetic data from HapMap 3 consisting of both resequencing and array-based genotyping data for the same set of individuals from multiple populations. We report that synthetic associations tend to be further away from the underlying risk alleles compared to "natural associations" (i.e. associations due to underlying common causal variants), but to a much lesser extent than previously predicted, with both the age and the effect size of the risk allele playing a part in this phenomenon. We find that while a synthetic association has a lower probability of capturing causal variants within its linkage disequilibrium block, sequencing around the associated variant need not extend substantially to have a high probability of capturing at least one causal variant. We also show that the minor allele frequency of synthetic associations is lower than of natural associations for most, but not all, loci that we explored. Finally, we find the variance in associated allele frequency to be a potential indicator of synthetic associations.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
Detection of common single nucleotide polymorphisms synthesizing quantitative trait association of rarer causal variants.Genome Res. 2011 Jul;21(7):1122-30. doi: 10.1101/gr.115832.110. Epub 2011 Mar 25. Genome Res. 2011. PMID: 21441355 Free PMC article.
-
Rare variants create synthetic genome-wide associations.PLoS Biol. 2010 Jan 26;8(1):e1000294. doi: 10.1371/journal.pbio.1000294. PLoS Biol. 2010. PMID: 20126254 Free PMC article.
-
Interpretation of association signals and identification of causal variants from genome-wide association studies.Am J Hum Genet. 2010 May 14;86(5):730-42. doi: 10.1016/j.ajhg.2010.04.003. Epub 2010 Apr 29. Am J Hum Genet. 2010. PMID: 20434130 Free PMC article.
-
Molecular genetic studies of complex phenotypes.Transl Res. 2012 Feb;159(2):64-79. doi: 10.1016/j.trsl.2011.08.001. Epub 2011 Aug 31. Transl Res. 2012. PMID: 22243791 Free PMC article. Review.
-
An evolving understanding of multiple causal variants underlying genetic association signals.Am J Hum Genet. 2025 Apr 3;112(4):741-750. doi: 10.1016/j.ajhg.2025.01.018. Epub 2025 Feb 17. Am J Hum Genet. 2025. PMID: 39965570 Review.
Cited by
-
High burden of private mutations due to explosive human population growth and purifying selection.BMC Genomics. 2014;15 Suppl 4(Suppl 4):S3. doi: 10.1186/1471-2164-15-S4-S3. Epub 2014 May 20. BMC Genomics. 2014. PMID: 25056720 Free PMC article.
-
Fine-mapping the HOXB region detects common variants tagging a rare coding allele: evidence for synthetic association in prostate cancer.PLoS Genet. 2014 Feb 13;10(2):e1004129. doi: 10.1371/journal.pgen.1004129. eCollection 2014 Feb. PLoS Genet. 2014. PMID: 24550738 Free PMC article.
-
Network analysis identifies protein clusters of functional importance in juvenile idiopathic arthritis.Arthritis Res Ther. 2014 May 8;16(3):R109. doi: 10.1186/ar4559. Arthritis Res Ther. 2014. PMID: 24886659 Free PMC article.
-
Whole-exome sequencing of 2,000 Danish individuals and the role of rare coding variants in type 2 diabetes.Am J Hum Genet. 2013 Dec 5;93(6):1072-86. doi: 10.1016/j.ajhg.2013.11.005. Epub 2013 Nov 27. Am J Hum Genet. 2013. PMID: 24290377 Free PMC article.
-
Deciphering the genetic architecture of low-penetrance susceptibility to colorectal cancer.Hum Mol Genet. 2013 Dec 15;22(24):5075-82. doi: 10.1093/hmg/ddt357. Epub 2013 Jul 30. Hum Mol Genet. 2013. PMID: 23904454 Free PMC article.
References
-
- Pritchard JK, Cox NJ. The allelic architecture of human disease genes: common disease-common variant…or not? Hum Mol Genet. 2002;11:2417–2423. - PubMed
-
- Reich DE, Lander ES. On the allelic spectrum of human disease. Trends Genet. 2001;17:502–510. - PubMed
-
- Frazer KA, Murray SS, Schork NJ, Topol EJ. Human genetic variation and its contribution to complex traits. Nat Rev Genet. 2009;10:241–251. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

