Proteomic profiling of the TRAF3 interactome network reveals a new role for the ER-to-Golgi transport compartments in innate immunity
- PMID: 22792062
- PMCID: PMC3390413
- DOI: 10.1371/journal.ppat.1002747
Proteomic profiling of the TRAF3 interactome network reveals a new role for the ER-to-Golgi transport compartments in innate immunity
Abstract
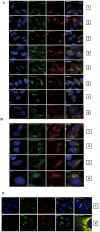
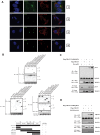
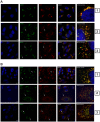
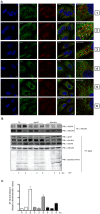
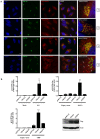
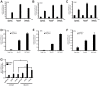
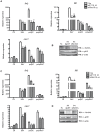
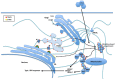
Tumor Necrosis Factor receptor-associated factor-3 (TRAF3) is a central mediator important for inducing type I interferon (IFN) production in response to intracellular double-stranded RNA (dsRNA). Here, we report the identification of Sec16A and p115, two proteins of the ER-to-Golgi vesicular transport system, as novel components of the TRAF3 interactome network. Notably, in non-infected cells, TRAF3 was found associated with markers of the ER-Exit-Sites (ERES), ER-to-Golgi intermediate compartment (ERGIC) and the cis-Golgi apparatus. Upon dsRNA and dsDNA sensing however, the Golgi apparatus fragmented into cytoplasmic punctated structures containing TRAF3 allowing its colocalization and interaction with Mitochondrial AntiViral Signaling (MAVS), the essential mitochondria-bound RIG-I-like Helicase (RLH) adaptor. In contrast, retention of TRAF3 at the ER-to-Golgi vesicular transport system blunted the ability of TRAF3 to interact with MAVS upon viral infection and consequently decreased type I IFN response. Moreover, depletion of Sec16A and p115 led to a drastic disorganization of the Golgi paralleled by the relocalization of TRAF3, which under these conditions was unable to associate with MAVS. Consequently, upon dsRNA and dsDNA sensing, ablation of Sec16A and p115 was found to inhibit IRF3 activation and anti-viral gene expression. Reciprocally, mild overexpression of Sec16A or p115 in Hec1B cells increased the activation of IFNβ, ISG56 and NF-κB -dependent promoters following viral infection and ectopic expression of MAVS and Tank-binding kinase-1 (TBK1). In line with these results, TRAF3 was found enriched in immunocomplexes composed of p115, Sec16A and TBK1 upon infection. Hence, we propose a model where dsDNA and dsRNA sensing induces the formation of membrane-bound compartments originating from the Golgi, which mediate the dynamic association of TRAF3 with MAVS leading to an optimal induction of innate immune responses.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Barbalat R, Ewald SE, Mouchess ML, Barton GM. Nucleic acid recognition by the innate immune system. Annu Rev Immunol. 2011;29:185–214. - PubMed
-
- Kato H, Sato S, Yoneyama M, Yamamoto M, Uematsu S, et al. Cell type-specific involvement of RIG-I in antiviral response. Immunity. 2005;23:19–28. - PubMed
-
- Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

