A regulatory loop involving PAX6, MITF, and WNT signaling controls retinal pigment epithelium development
- PMID: 22792072
- PMCID: PMC3390378
- DOI: 10.1371/journal.pgen.1002757
A regulatory loop involving PAX6, MITF, and WNT signaling controls retinal pigment epithelium development
Abstract
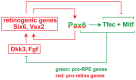
The separation of the optic neuroepithelium into future retina and retinal pigment epithelium (RPE) is a critical event in early eye development in vertebrates. Here we show in mice that the transcription factor PAX6, well-known for its retina-promoting activity, also plays a crucial role in early pigment epithelium development. This role is seen, however, only in a background genetically sensitized by mutations in the pigment cell transcription factor MITF. In fact, a reduction in Pax6 gene dose exacerbates the RPE-to-retina transdifferentiation seen in embryos homozygous for an Mitf null allele, and it induces such a transdifferentiation in embryos that are either heterozygous for the Mitf null allele or homozygous for an RPE-specific hypomorphic Mitf allele generated by targeted mutation. Conversely, an increase in Pax6 gene dose interferes with transdifferentiation even in homozygous Mitf null embryos. Gene expression analyses show that, together with MITF or its paralog TFEC, PAX6 suppresses the expression of Fgf15 and Dkk3. Explant culture experiments indicate that a combination of FGF and DKK3 promote retina formation by inhibiting canonical WNT signaling and stimulating the expression of retinogenic genes, including Six6 and Vsx2. Our results demonstrate that in conjunction with Mitf/Tfec Pax6 acts as an anti-retinogenic factor, whereas in conjunction with retinogenic genes it acts as a pro-retinogenic factor. The results suggest that careful manipulation of the Pax6 regulatory circuit may facilitate the generation of retinal and pigment epithelium cells from embryonic or induced pluripotent stem cells.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
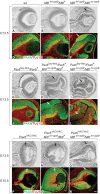
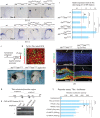

 , non-consensus
, non-consensus  ), with positions indicated relative to translation start sites. (P) ChIP assays performed with indicated antibodies on RPE fractions dissected from E11.5 wild-type embryonic eyes. Amplicons as indicated in (O). (Q) ChIP assays performed with the indicated antibodies on optic vesicle tissue dissected from E10.5 embryonic eyes of the indicated genotypes. Anti-acetyl H3 signal represents active chromatin domains and anti-dimethyl H3K9 signal inactive chromatin domains. For details see text.
), with positions indicated relative to translation start sites. (P) ChIP assays performed with indicated antibodies on RPE fractions dissected from E11.5 wild-type embryonic eyes. Amplicons as indicated in (O). (Q) ChIP assays performed with the indicated antibodies on optic vesicle tissue dissected from E10.5 embryonic eyes of the indicated genotypes. Anti-acetyl H3 signal represents active chromatin domains and anti-dimethyl H3K9 signal inactive chromatin domains. For details see text.
Comment in
-
Both PAX6 and MITF are required for pigment epithelium development in vivo.Pigment Cell Melanoma Res. 2012 Sep;25(5):541-3. doi: 10.1111/j.1755-148x.2012.01037.x. Pigment Cell Melanoma Res. 2012. PMID: 23094276 No abstract available.
References
-
- Miller SS, Maminishkis A, Li R, Adijanto J. Retinal Pigment Epithelium: Cytokine Modulation of Epithelial Physiology. In: Dartt DA, editor. Encyclopedia of the Eye: Oxford University. Academic Press; 2010. pp. 89–100. editor.
-
- Hemesath TJ, Steingrimsson E, McGill G, Hansen MJ, Vaught J, et al. microphthalmia, a critical factor in melanocyte development, defines a discrete transcription factor family. Genes Dev. 1994;8:2770–2780. - PubMed
-
- Hodgkinson CA, Moore KJ, Nakayama A, Steingrimsson E, Copeland NG, et al. Mutations at the mouse microphthalmia locus are associated with defects in a gene encoding a novel basic-helix-loop-helix-zipper protein. Cell. 1993;74:395–404. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

