Comparative genomics of plant-associated Pseudomonas spp.: insights into diversity and inheritance of traits involved in multitrophic interactions
- PMID: 22792073
- PMCID: PMC3390384
- DOI: 10.1371/journal.pgen.1002784
Comparative genomics of plant-associated Pseudomonas spp.: insights into diversity and inheritance of traits involved in multitrophic interactions
Abstract
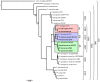
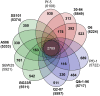
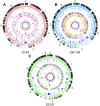
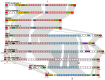
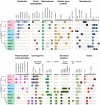
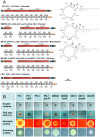
We provide here a comparative genome analysis of ten strains within the Pseudomonas fluorescens group including seven new genomic sequences. These strains exhibit a diverse spectrum of traits involved in biological control and other multitrophic interactions with plants, microbes, and insects. Multilocus sequence analysis placed the strains in three sub-clades, which was reinforced by high levels of synteny, size of core genomes, and relatedness of orthologous genes between strains within a sub-clade. The heterogeneity of the P. fluorescens group was reflected in the large size of its pan-genome, which makes up approximately 54% of the pan-genome of the genus as a whole, and a core genome representing only 45-52% of the genome of any individual strain. We discovered genes for traits that were not known previously in the strains, including genes for the biosynthesis of the siderophores achromobactin and pseudomonine and the antibiotic 2-hexyl-5-propyl-alkylresorcinol; novel bacteriocins; type II, III, and VI secretion systems; and insect toxins. Certain gene clusters, such as those for two type III secretion systems, are present only in specific sub-clades, suggesting vertical inheritance. Almost all of the genes associated with multitrophic interactions map to genomic regions present in only a subset of the strains or unique to a specific strain. To explore the evolutionary origin of these genes, we mapped their distributions relative to the locations of mobile genetic elements and repetitive extragenic palindromic (REP) elements in each genome. The mobile genetic elements and many strain-specific genes fall into regions devoid of REP elements (i.e., REP deserts) and regions displaying atypical tri-nucleotide composition, possibly indicating relatively recent acquisition of these loci. Collectively, the results of this study highlight the enormous heterogeneity of the P. fluorescens group and the importance of the variable genome in tailoring individual strains to their specific lifestyles and functional repertoire.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Lessie TG, Phibbs PV., Jr Alternative pathways of carbohydrate utilization in Pseudomonads. Annu Rev Microbiol. 1984;38:359–388. - PubMed
-
- Gross H, Loper JE. Genomics of secondary metabolite production by Pseudomonas spp. Nat Prod Rep. 2009;26:1408–1446. - PubMed
-
- Raaijmakers JM, Vlami M, de Souza JT. Antibiotic production by bacterial biocontrol agents. Antonie Van Leeuwenhoek. 2002;81:537–547. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

