Gene conversion occurs within the mating-type locus of Cryptococcus neoformans during sexual reproduction
- PMID: 22792079
- PMCID: PMC3390403
- DOI: 10.1371/journal.pgen.1002810
Gene conversion occurs within the mating-type locus of Cryptococcus neoformans during sexual reproduction
Abstract
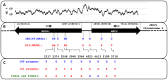
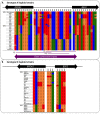
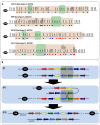
Meiotic recombination of sex chromosomes is thought to be repressed in organisms with heterogametic sex determination (e.g. mammalian X/Y chromosomes), due to extensive divergence and chromosomal rearrangements between the two chromosomes. However, proper segregation of sex chromosomes during meiosis requires crossing-over occurring within the pseudoautosomal regions (PAR). Recent studies reveal that recombination, in the form of gene conversion, is widely distributed within and may have played important roles in the evolution of some chromosomal regions within which recombination was thought to be repressed, such as the centromere cores of maize. Cryptococcus neoformans, a major human pathogenic fungus, has an unusually large mating-type locus (MAT, >100 kb), and the MAT alleles from the two opposite mating-types show extensive nucleotide sequence divergence and chromosomal rearrangements, mirroring characteristics of sex chromosomes. Meiotic recombination was assumed to be repressed within the C. neoformans MAT locus. A previous study identified recombination hot spots flanking the C. neoformans MAT, and these hot spots are associated with high GC content. Here, we investigated a GC-rich intergenic region located within the MAT locus of C. neoformans to establish if this region also exhibits unique recombination behavior during meiosis. Population genetics analysis of natural C. neoformans isolates revealed signals of homogenization spanning this GC-rich intergenic region within different C. neoformans lineages, consistent with a model in which gene conversion of this region during meiosis prevents it from diversifying within each lineage. By analyzing meiotic progeny from laboratory crosses, we found that meiotic recombination (gene conversion) occurs around the GC-rich intergenic region at a frequency equal to or greater than the meiotic recombination frequency observed in other genomic regions. We discuss the implications of these findings with regards to the possible functional and evolutionary importance of gene conversion within the C. neoformans MAT locus and, more generally, in fungi.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures






References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

