Over expression of wild type or a catalytically dead mutant of Sirtuin 6 does not influence NFκB responses
- PMID: 22792191
- PMCID: PMC3391194
- DOI: 10.1371/journal.pone.0039847
Over expression of wild type or a catalytically dead mutant of Sirtuin 6 does not influence NFκB responses
Erratum in
- PLoS One. 2012;7(9). doi:10.1371/annotation/9c1a5765-6471-401f-bc74-a916c8ead46f
Abstract
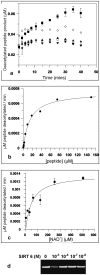
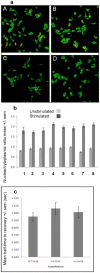
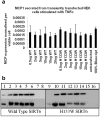
SIRT6 is involved in inflammation, aging and metabolism potentially by modulating the functions of both NFκB and HIF1α. Since it is possible to make small molecule activators and inhibitors of Sirtuins we wished to establish biochemical and cellular assays both to assist in drug discovery efforts and to validate whether SIRT6 represents a valid drug target for these indications. We confirmed in cellular assays that SIRT6 can deacetylate acetylated-histone H3 lysine 9 (H3K9Ac), however this deacetylase activity is unusually low in biochemical assays. In an effort to develop alternative assay formats we observed that SIRT6 overexpression had no influence on TNFα induced nuclear translocation of NFκB, nor did it have an effect on nuclear mobility of RelA/p65. In an effort to identify a gene expression profile that could be used to identify a SIRT6 readout we conducted genome-wide expression studies. We observed that overexpression of SIRT6 had little influence on NFκB-dependent genes, but overexpression of the catalytically inactive mutant affected gene expression in developmental pathways.
Conflict of interest statement
Figures




Similar articles
-
SIRT6 links histone H3 lysine 9 deacetylation to NF-kappaB-dependent gene expression and organismal life span.Cell. 2009 Jan 9;136(1):62-74. doi: 10.1016/j.cell.2008.10.052. Cell. 2009. PMID: 19135889 Free PMC article.
-
Sirtuin 6 inhibits myofibroblast differentiation via inactivating transforming growth factor-β1/Smad2 and nuclear factor-κB signaling pathways in human fetal lung fibroblasts.J Cell Biochem. 2019 Jan;120(1):93-104. doi: 10.1002/jcb.27128. Epub 2018 Sep 19. J Cell Biochem. 2019. PMID: 30230565
-
Sirtuin 6 protects cardiomyocytes from hypertrophy in vitro via inhibition of NF-κB-dependent transcriptional activity.Br J Pharmacol. 2013 Jan;168(1):117-28. doi: 10.1111/j.1476-5381.2012.01903.x. Br J Pharmacol. 2013. PMID: 22335191 Free PMC article.
-
Emerging roles of SIRT6 on telomere maintenance, DNA repair, metabolism and mammalian aging.Mol Cell Biochem. 2012 May;364(1-2):345-50. doi: 10.1007/s11010-012-1236-8. Mol Cell Biochem. 2012. PMID: 22286818 Review.
-
The Role of Sirtuin 6 in the Deacetylation of Histone Proteins as a Factor in the Progression of Neoplastic Disease.Int J Mol Sci. 2023 Dec 29;25(1):497. doi: 10.3390/ijms25010497. Int J Mol Sci. 2023. PMID: 38203666 Free PMC article. Review.
Cited by
-
SIRT6 delays cellular senescence by promoting p27Kip1 ubiquitin-proteasome degradation.Aging (Albany NY). 2016 Sep 16;8(10):2308-2323. doi: 10.18632/aging.101038. Aging (Albany NY). 2016. PMID: 27794562 Free PMC article.
-
ModuleBlast: identifying activated sub-networks within and across species.Nucleic Acids Res. 2015 Feb 18;43(3):e20. doi: 10.1093/nar/gku1224. Epub 2014 Nov 26. Nucleic Acids Res. 2015. PMID: 25428368 Free PMC article.
-
Biochemical characterization of sirtuin 6 in the brain and its involvement in oxidative stress response.Neurochem Res. 2015 Jan;40(1):59-69. doi: 10.1007/s11064-014-1465-1. Epub 2014 Nov 1. Neurochem Res. 2015. PMID: 25366464
-
Mammalian target of rapamycin complex 2 (mTORC2) controls glycolytic gene expression by regulating Histone H3 Lysine 56 acetylation.Cell Cycle. 2018;17(1):110-123. doi: 10.1080/15384101.2017.1404207. Epub 2018 Jan 8. Cell Cycle. 2018. PMID: 29143563 Free PMC article.
-
SIRT6 overexpression in the nucleus protects mouse retinal pigment epithelium from oxidative stress.Life Sci Alliance. 2023 Apr 25;6(7):e202201448. doi: 10.26508/lsa.202201448. Print 2023 Jul. Life Sci Alliance. 2023. PMID: 37185874 Free PMC article.
References
-
- Lavu S, Boss O, Elliott PJ, Lambert PD. Sirtuins-novel therapeutic targets to treat age-associated diseases. Nat Rev Drug Discov. 2008;7:841–853. - PubMed
-
- Frye RA. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem Biophys Res Commun. 2000;273:793–798. - PubMed
-
- Liszt G, Ford E, Kurtev M, Guarente L. Mouse Sir2 homolog SIRT6 is a nuclear ADP-ribosyltransferase. J Biol Chem. 2005;280:21313–21320. - PubMed
-
- Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124:315–329. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

