Transcript profile of the response of two soybean genotypes to potassium deficiency
- PMID: 22792192
- PMCID: PMC3390323
- DOI: 10.1371/journal.pone.0039856
Transcript profile of the response of two soybean genotypes to potassium deficiency
Abstract
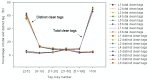
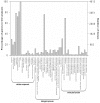
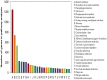
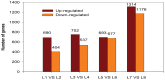
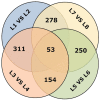
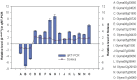
The macronutrient potassium (K) is essential to plant growth and development. Crop yield potential is often affected by lack of soluble K. The molecular regulation mechanism of physiological and biochemical responses to K starvation in soybean roots and shoots is not fully understood. In the present study, two soybean varieties were subjected to low-K stress conditions: a low-K-tolerant variety (You06-71) and a low-K-sensitive variety (HengChun04-11). Eight libraries were generated for analysis: 2 genotypes ×2 tissues (roots and shoots) ×2 time periods [short term (0.5 to 12 h) and long term (3 to 12 d)]. RNA derived from the roots and shoots of these two varieties across two periods (short term and long term) were sequenced and the transcriptomes were compared using high-throughput tag-sequencing. To this end, a large number of clean tags (tags used for analysis after removal of dirty tags) corresponding to distinct tags (all types of clean tags) were identified in eight libraries (L1, You06-71-root short term; L2, HengChun04-11-root short term; L3, You06-71-shoot short term; L4, HengChun04-11-shoot short term; L5, You06-71-root long term; L6, HengChun04-11-root long term; L7, You06-71-shoot long term; L8, HengChun04-11-shoot long term). All clean tags were mapped to the available soybean (Glycine max) transcript database (http://www.soybase.org). Many genes showed substantial differences in expression across the libraries. In total, 5,440 transcripts involved in 118 KEGG pathways were either up- or down-regulated. Fifteen genes were randomly selected and their expression levels were confirmed using quantitative RT-PCR. Our results provide preliminary information on the molecular mechanism of potassium absorption and transport under low-K stress conditions in different soybean tissues.
Conflict of interest statement
Figures

References
-
- Adams F. Soil solution. The plant root and its environment Univ Press of Virginia, Charlottesville. 1974. pp. 441–481.
-
- Tisdale S, Nelson W, Beaton J, Havlin J. Soil and fertilizer nitrogen. Soil fertility and fertilizers. 1993;5:109–175.
-
- Schroeder JI, Kwak JM, Allen GJ. Guard cell abscisic acid signalling and engineering drought hardiness in plants. Nature. 2001;410:327–330. - PubMed
-
- Assmann SM. Open Stomatal opens the door to ABA signaling in Arabidopsis guard cells. Trends Plant Sci. 2003;8:151–153. - PubMed
-
- Sutter JU, Sieben C, Hartel A, Eisenach C, Thiel G, et al. Abscisic acid triggers the endocytosis of the Arabidopsis KAT1 K+ channel and its recycling to the plasma membrane. Curr Biol. 2007;17:1396–1402. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

