A versatile method for cell-specific profiling of translated mRNAs in Drosophila
- PMID: 22792260
- PMCID: PMC3391276
- DOI: 10.1371/journal.pone.0040276
A versatile method for cell-specific profiling of translated mRNAs in Drosophila
Erratum in
-
Correction: A Versatile Method for Cell-Specific Profiling of Translated mRNAs in Drosophila.PLoS One. 2013 Jul 24;8(7):10.1371/annotation/39194a57-4480-4f8e-b6fa-e7e0993d029b. doi: 10.1371/annotation/39194a57-4480-4f8e-b6fa-e7e0993d029b. Print 2013. PLoS One. 2013. PMID: 23894271 Free PMC article.
Abstract
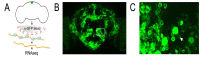
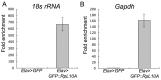
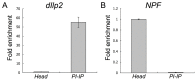
In Drosophila melanogaster few methods exist to perform rapid cell-type or tissue-specific expression profiling. A translating ribosome affinity purification (TRAP) method to profile actively translated mRNAs has been developed for use in a number of multicellular organisms although it has only been implemented to examine limited sets of cell- or tissue-types in these organisms. We have adapted the TRAP method for use in the versatile GAL4/UAS system of Drosophila allowing profiling of almost any tissue/cell-type with a single genetic cross. We created transgenic strains expressing a GFP-tagged ribosomal protein, RpL10A, under the control of the UAS promoter to perform cell-type specific translatome profiling. The GFP::RpL10A fusion protein incorporates efficiently into ribosomes and polysomes. Polysome affinity purification strongly enriches mRNAs from expected genes in the targeted tissues with sufficient sensitivity to analyze expression in small cell populations. This method can be used to determine the unique translatome profiles in different cell-types under varied physiological, pharmacological and pathological conditions.
Conflict of interest statement
Figures


Similar articles
-
TRAP-rc, Translating Ribosome Affinity Purification from Rare Cell Populations of Drosophila Embryos.J Vis Exp. 2015 Sep 10;(103):52985. doi: 10.3791/52985. J Vis Exp. 2015. PMID: 26381166 Free PMC article.
-
A split green fluorescent protein system to enhance spatial and temporal sensitivity of translating ribosome affinity purification.Plant J. 2022 Jul;111(1):304-315. doi: 10.1111/tpj.15779. Epub 2022 May 10. Plant J. 2022. PMID: 35436375 Free PMC article.
-
Development of translating ribosome affinity purification for zebrafish.Genesis. 2013 Mar;51(3):187-92. doi: 10.1002/dvg.22363. Epub 2013 Feb 26. Genesis. 2013. PMID: 23281262 Free PMC article.
-
Translatome profiling: methods for genome-scale analysis of mRNA translation.Brief Funct Genomics. 2016 Jan;15(1):22-31. doi: 10.1093/bfgp/elu045. Epub 2014 Nov 6. Brief Funct Genomics. 2016. PMID: 25380596 Review.
-
Purification, identification, and functional analysis of polysomes from the human pathogen Staphylococcus aureus.Methods. 2017 Mar 15;117:59-66. doi: 10.1016/j.ymeth.2016.10.003. Epub 2016 Oct 8. Methods. 2017. PMID: 27729294 Review.
Cited by
-
The Zinc-BED Transcription Factor Bedwarfed Promotes Proportional Dendritic Growth and Branching through Transcriptional and Translational Regulation in Drosophila.Int J Mol Sci. 2023 Mar 28;24(7):6344. doi: 10.3390/ijms24076344. Int J Mol Sci. 2023. PMID: 37047316 Free PMC article.
-
Simultaneous Transcriptional and Epigenomic Profiling from Specific Cell Types within Heterogeneous Tissues In Vivo.Cell Rep. 2017 Jan 24;18(4):1048-1061. doi: 10.1016/j.celrep.2016.12.087. Cell Rep. 2017. PMID: 28122230 Free PMC article.
-
Development of a tissue-specific ribosome profiling approach in Drosophila enables genome-wide evaluation of translational adaptations.PLoS Genet. 2017 Dec 1;13(12):e1007117. doi: 10.1371/journal.pgen.1007117. eCollection 2017 Dec. PLoS Genet. 2017. PMID: 29194454 Free PMC article.
-
Translational regulation enhances distinction of cell types in the nervous system.Elife. 2024 Jul 16;12:RP90713. doi: 10.7554/eLife.90713. Elife. 2024. PMID: 39010741 Free PMC article.
-
Temporally and spatially restricted gene expression profiling.Curr Genomics. 2014 Aug;15(4):278-92. doi: 10.2174/1389202915666140602230106. Curr Genomics. 2014. PMID: 25132798 Free PMC article.
References
-
- Venken KJ, Bellen HJ. Transgenesis upgrades for Drosophila melanogaster. Development. 2007;134:3571–84. - PubMed
-
- Jones WD. The expanding reach of the GAL4/UAS system into the behavioral neurobiology of Drosophila. BMB Rep. 2009;42:705–12. - PubMed
-
- Chintapalli VR, Wang J, Dow JA. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat Genet. 2007;39:715–20. - PubMed
-
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

