POWRS: position-sensitive motif discovery
- PMID: 22792292
- PMCID: PMC3390389
- DOI: 10.1371/journal.pone.0040373
POWRS: position-sensitive motif discovery
Abstract
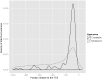
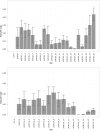
Transcription factors and the short, often degenerate DNA sequences they recognize are central regulators of gene expression, but their regulatory code is challenging to dissect experimentally. Thus, computational approaches have long been used to identify putative regulatory elements from the patterns in promoter sequences. Here we present a new algorithm "POWRS" (POsition-sensitive WoRd Set) for identifying regulatory sequence motifs, specifically developed to address two common shortcomings of existing algorithms. First, POWRS uses the position-specific enrichment of regulatory elements near transcription start sites to significantly increase sensitivity, while providing new information about the preferred localization of those elements. Second, POWRS forgoes position weight matrices for a discrete motif representation that appears more resistant to over-generalization. We apply this algorithm to discover sequences related to constitutive, high-level gene expression in the model plant Arabidopsis thaliana, and then experimentally validate the importance of those elements by systematically mutating two endogenous promoters and measuring the effect on gene expression levels. This provides a foundation for future efforts to rationally engineer gene expression in plants, a problem of great importance in developing biotech crop varieties.
Availability: BSD-licensed Python code at http://grassrootsbio.com/papers/powrs/.
Conflict of interest statement
Figures

Similar articles
-
Paired-end analysis of transcription start sites in Arabidopsis reveals plant-specific promoter signatures.Plant Cell. 2014 Jul;26(7):2746-60. doi: 10.1105/tpc.114.125617. Epub 2014 Jul 17. Plant Cell. 2014. PMID: 25035402 Free PMC article.
-
Athena: a resource for rapid visualization and systematic analysis of Arabidopsis promoter sequences.Bioinformatics. 2005 Dec 15;21(24):4411-3. doi: 10.1093/bioinformatics/bti714. Epub 2005 Oct 13. Bioinformatics. 2005. PMID: 16223790
-
De-novo discovery of differentially abundant transcription factor binding sites including their positional preference.PLoS Comput Biol. 2011 Feb 10;7(2):e1001070. doi: 10.1371/journal.pcbi.1001070. PLoS Comput Biol. 2011. PMID: 21347314 Free PMC article.
-
BLSSpeller: exhaustive comparative discovery of conserved cis-regulatory elements.Bioinformatics. 2015 Dec 1;31(23):3758-66. doi: 10.1093/bioinformatics/btv466. Epub 2015 Aug 8. Bioinformatics. 2015. PMID: 26254488 Free PMC article.
-
Bounded search for de novo identification of degenerate cis-regulatory elements.BMC Bioinformatics. 2006 May 15;7:254. doi: 10.1186/1471-2105-7-254. BMC Bioinformatics. 2006. PMID: 16700920 Free PMC article.
Cited by
-
Computationally derived RNA polymerase III promoters enable maize genome editing.Front Plant Sci. 2025 Mar 19;16:1540425. doi: 10.3389/fpls.2025.1540425. eCollection 2025. Front Plant Sci. 2025. PMID: 40177017 Free PMC article.
-
Conserved plant transcriptional responses to microgravity from two consecutive spaceflight experiments.Front Plant Sci. 2024 Jan 8;14:1308713. doi: 10.3389/fpls.2023.1308713. eCollection 2023. Front Plant Sci. 2024. PMID: 38259952 Free PMC article.
-
Dempster-Shafer Theory for the Prediction of Auxin-Response Elements (AuxREs) in Plant Genomes.Biomed Res Int. 2018 Nov 1;2018:3837060. doi: 10.1155/2018/3837060. eCollection 2018. Biomed Res Int. 2018. PMID: 30515394 Free PMC article.
-
Expression Elements Derived From Plant Sequences Provide Effective Gene Expression Regulation and New Opportunities for Plant Biotechnology Traits.Front Plant Sci. 2021 Oct 22;12:712179. doi: 10.3389/fpls.2021.712179. eCollection 2021. Front Plant Sci. 2021. PMID: 34745155 Free PMC article.
-
Discriminative motif optimization based on perceptron training.Bioinformatics. 2014 Apr 1;30(7):941-8. doi: 10.1093/bioinformatics/btt748. Epub 2013 Dec 24. Bioinformatics. 2014. PMID: 24369152 Free PMC article.
References
-
- Sun HQ, Low MY, Hsu WJ, Tan CW, Rajapakse JC. Tree-structured algorithm for long weak motif discovery. Bioinformatics. 2011;27:2641–2647. - PubMed
-
- Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

