A degenerate primer MOB typing (DPMT) method to classify gamma-proteobacterial plasmids in clinical and environmental settings
- PMID: 22792321
- PMCID: PMC3394729
- DOI: 10.1371/journal.pone.0040438
A degenerate primer MOB typing (DPMT) method to classify gamma-proteobacterial plasmids in clinical and environmental settings
Abstract
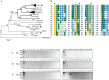
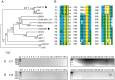
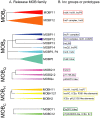
Transmissible plasmids are responsible for the spread of genetic determinants, such as antibiotic resistance or virulence traits, causing a large ecological and epidemiological impact. Transmissible plasmids, either conjugative or mobilizable, have in common the presence of a relaxase gene. Relaxases were previously classified in six protein families according to their phylogeny. Degenerate primers hybridizing to coding sequences of conserved amino acid motifs were designed to amplify related relaxase genes from γ-Proteobacterial plasmids. Specificity and sensitivity of a selected set of 19 primer pairs were first tested using a collection of 33 reference relaxases, representing the diversity of γ-Proteobacterial plasmids. The validated set was then applied to the analysis of two plasmid collections obtained from clinical isolates. The relaxase screening method, which we call "Degenerate Primer MOB Typing" or DPMT, detected not only most known Inc/Rep groups, but also a plethora of plasmids not previously assigned to any Inc group or Rep-type.
Conflict of interest statement
Figures





Similar articles
-
Degenerate primer MOB typing of multiresistant clinical isolates of E. coli uncovers new plasmid backbones.Plasmid. 2015 Jan;77:17-27. doi: 10.1016/j.plasmid.2014.11.003. Epub 2014 Nov 22. Plasmid. 2015. PMID: 25463772
-
The diversity of conjugative relaxases and its application in plasmid classification.FEMS Microbiol Rev. 2009 May;33(3):657-87. doi: 10.1111/j.1574-6976.2009.00168.x. FEMS Microbiol Rev. 2009. PMID: 19396961 Review.
-
Plasmid Typing and Classification.Methods Mol Biol. 2020;2075:309-321. doi: 10.1007/978-1-4939-9877-7_22. Methods Mol Biol. 2020. PMID: 31584172
-
PLASmid TAXonomic PCR (PlasTax-PCR), a Multiplex Relaxase MOB Typing to Assort Plasmids into Taxonomic Units.Methods Mol Biol. 2022;2392:127-142. doi: 10.1007/978-1-0716-1799-1_10. Methods Mol Biol. 2022. PMID: 34773620
-
A classification scheme for mobilization regions of bacterial plasmids.FEMS Microbiol Rev. 2004 Feb;28(1):79-100. doi: 10.1016/j.femsre.2003.09.001. FEMS Microbiol Rev. 2004. PMID: 14975531 Review.
Cited by
-
ExplorePipolin: reconstruction and annotation of piPolB-encoding bacterial mobile elements from draft genomes.Bioinform Adv. 2022 Aug 10;2(1):vbac056. doi: 10.1093/bioadv/vbac056. eCollection 2022. Bioinform Adv. 2022. PMID: 36699382 Free PMC article.
-
Approaches for characterizing and tracking hospital-associated multidrug-resistant bacteria.Cell Mol Life Sci. 2021 Mar;78(6):2585-2606. doi: 10.1007/s00018-020-03717-2. Epub 2021 Feb 13. Cell Mol Life Sci. 2021. PMID: 33582841 Free PMC article. Review.
-
Molecular Analysis of Carbapenem and Aminoglycoside Resistance Genes in Carbapenem-Resistant Pseudomonas aeruginosa Clinical Strains: A Challenge for Tertiary Care Hospitals.Antibiotics (Basel). 2024 Feb 16;13(2):191. doi: 10.3390/antibiotics13020191. Antibiotics (Basel). 2024. PMID: 38391577 Free PMC article.
-
A global survey of Salmonella plasmids and their associations with antimicrobial resistance.Microb Genom. 2023 May;9(5):mgen001002. doi: 10.1099/mgen.0.001002. Microb Genom. 2023. PMID: 37200081 Free PMC article.
-
Novel Pseudomonas aeruginosa Strains Co-Harbouring bla NDM-1 Metallo β-Lactamase and mcr-1 Isolated from Immunocompromised Paediatric Patients.Infect Drug Resist. 2022 Jun 8;15:2929-2936. doi: 10.2147/IDR.S368566. eCollection 2022. Infect Drug Resist. 2022. PMID: 35706928 Free PMC article.
References
-
- de la Cruz F, Davies J. Horizontal gene transfer and the origin of species: lessons from bacteria. Trends Microbiol. 2000;8:128–133. - PubMed
-
- Garcillan-Barcia MP, Alvarado A, de la Cruz F. Identification of bacterial plasmids based on mobility and plasmid population biology. FEMS Microbiol Rev. 2011;35:936–956. - PubMed
-
- Austin S, Nordstrom K. Partition-mediated incompatibility of bacterial plasmids. Cell. 1990;60:351–354. - PubMed

