A new Cre driver mouse line, Tcf21/Pod1-Cre, targets metanephric mesenchyme
- PMID: 22792366
- PMCID: PMC3391250
- DOI: 10.1371/journal.pone.0040547
A new Cre driver mouse line, Tcf21/Pod1-Cre, targets metanephric mesenchyme
Abstract
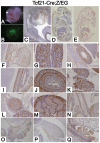
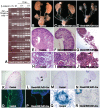
Conditional gene targeting in mice has provided great insight into the role of gene function in kidney development and disease. Although a number of Cre-driver mouse strains already exist for the kidney, development of additional strains with unique expression patterns is needed. Here we report the generation and validation of a Tcf21/Pod1-Cre driver strain that expresses Cre recombinase throughout the condensing and stromal mesenchyme of developing kidneys and in their derivatives including epithelial components of the nephron and interstitial cells. To test the efficiency of this line, we crossed it to mice transgenic for either loss or gain of function β-catenin conditional alleles. Mice with deletion of β-catenin from Tcf21-expressing cells are born with hypoplastic kidneys, hydroureters and hydronephrosis. By contrast, Tcf21-Cre driven gain of function for β-catenin in mice results in fused midline kidneys and hypoplastic kidneys. Finally, we report the first renal mesenchymal deletion of Patched1 (Ptch1), the receptor for sonic hedgehog (Shh), which results in renal cysts demonstrating a functional role of Shh signaling pathway in renal cystogensis. In summary, we report the generation and validation of a new Cre driver strain that provides robust excision in metanephric mesenchyme.
Conflict of interest statement
Figures



Similar articles
-
Transcription Factor 21 Is Required for Branching Morphogenesis and Regulates the Gdnf-Axis in Kidney Development.J Am Soc Nephrol. 2018 Dec;29(12):2795-2808. doi: 10.1681/ASN.2017121278. Epub 2018 Oct 30. J Am Soc Nephrol. 2018. PMID: 30377232 Free PMC article.
-
Efficient inducible Cre-mediated recombination in Tcf21 cell lineages in the heart and kidney.Genesis. 2011 Nov;49(11):870-7. doi: 10.1002/dvg.20750. Epub 2011 Aug 18. Genesis. 2011. PMID: 21432986 Free PMC article.
-
A mouse line expressing Sall1-driven inducible Cre recombinase in the kidney mesenchyme.Genesis. 2010 Mar;48(3):207-12. doi: 10.1002/dvg.20603. Genesis. 2010. PMID: 20127799
-
Conditional mesenchymal disruption of pkd1 results in osteopenia and polycystic kidney disease.PLoS One. 2012;7(9):e46038. doi: 10.1371/journal.pone.0046038. Epub 2012 Sep 21. PLoS One. 2012. PMID: 23029375 Free PMC article.
-
Defining the genetic blueprint of kidney development.Pediatr Nephrol. 2011 Sep;26(9):1469-78. doi: 10.1007/s00467-011-1807-z. Epub 2011 Feb 19. Pediatr Nephrol. 2011. PMID: 21336810 Review.
Cited by
-
A prioritization tool for cilia-associated genes and their in vivo resources unveils new avenues for ciliopathy research.Dis Model Mech. 2024 Oct 1;17(10):dmm052000. doi: 10.1242/dmm.052000. Epub 2024 Oct 14. Dis Model Mech. 2024. PMID: 39263856 Free PMC article.
-
The Good and Bad of β-Catenin in Kidney Development and Renal Dysplasia.Front Cell Dev Biol. 2015 Dec 22;3:81. doi: 10.3389/fcell.2015.00081. eCollection 2015. Front Cell Dev Biol. 2015. PMID: 26734608 Free PMC article. Review.
-
Generation of the podocyte and tubular components of an amniote kidney: timing of specification and a role for Wnt signaling.Development. 2013 Nov;140(22):4565-73. doi: 10.1242/dev.097063. Epub 2013 Oct 23. Development. 2013. PMID: 24154527 Free PMC article.
-
Single-cell analysis of mesenchymal cells in permeable neural vasculature reveals novel diverse subpopulations of fibroblasts.Fluids Barriers CNS. 2024 Apr 5;21(1):31. doi: 10.1186/s12987-024-00535-7. Fluids Barriers CNS. 2024. PMID: 38575991 Free PMC article.
-
Genetic tools for identifying and manipulating fibroblasts in the mouse.Differentiation. 2016 Sep;92(3):66-83. doi: 10.1016/j.diff.2016.05.009. Epub 2016 Jun 21. Differentiation. 2016. PMID: 27342817 Free PMC article. Review.
References
-
- Maezawa Y, Kreidberg JA, Quaggin SE. CHAPTER 1- Embryology of the Kidney. Brenner and Rector's the Kidney. Philadelphia, PA: W.B. Saunders Company. 2012.
-
- Srinivas S, Goldberg MR, Watanabe T, D'Agati V, al-Awqati Q, et al. Expression of green fluorescent protein in the ureteric bud of transgenic mice: a new tool for the analysis of ureteric bud morphogenesis. Dev Genet. 1999;24:241–251. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

