Identification of a polyketide synthase required for alternariol (AOH) and alternariol-9-methyl ether (AME) formation in Alternaria alternata
- PMID: 22792370
- PMCID: PMC3391263
- DOI: 10.1371/journal.pone.0040564
Identification of a polyketide synthase required for alternariol (AOH) and alternariol-9-methyl ether (AME) formation in Alternaria alternata
Abstract
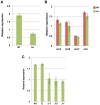
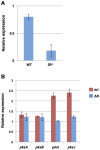
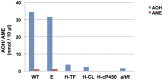
Alternaria alternata produces more than 60 secondary metabolites, among which alternariol (AOH) and alternariol-9-methyl ether (AME) are important mycotoxins. Whereas the toxicology of these two polyketide-based compounds has been studied, nothing is known about the genetics of their biosynthesis. One of the postulated core enzymes in the biosynthesis of AOH and AME is polyketide synthase (PKS). In a draft genome sequence of A. alternata we identified 10 putative PKS-encoding genes. The timing of the expression of two PKS genes, pksJ and pksH, correlated with the production of AOH and AME. The PksJ and PksH proteins are predicted to be 2222 and 2821 amino acids in length, respectively. They are both iterative type I reducing polyketide synthases. PksJ harbors a peroxisomal targeting sequence at the C-terminus, suggesting that the biosynthesis occurs at least partly in these organelles. In the vicinity of pksJ we found a transcriptional regulator, altR, involved in pksJ induction and a putative methyl transferase, possibly responsible for AME formation. Downregulation of pksJ and altR caused a large decrease of alternariol formation, suggesting that PksJ is the polyketide synthase required for the postulated Claisen condensations during the biosynthesis. No other enzymes appeared to be required. PksH downregulation affected pksJ expression and thus caused an indirect effect on AOH production.
Conflict of interest statement
Figures






Similar articles
-
Alternariol as virulence and colonization factor of Alternaria alternata during plant infection.Mol Microbiol. 2019 Jul;112(1):131-146. doi: 10.1111/mmi.14258. Epub 2019 Apr 23. Mol Microbiol. 2019. PMID: 30947377
-
Genome-Wide Analysis of Microsatellites in Alternaria arborescens and Elucidation of the Function of Polyketide Synthase (PksJ).Interdiscip Sci. 2018 Dec;10(4):813-822. doi: 10.1007/s12539-017-0251-6. Epub 2017 Oct 3. Interdiscip Sci. 2018. PMID: 28975513
-
LaeA and VeA are involved in growth morphology, asexual development, and mycotoxin production in Alternaria alternata.Int J Food Microbiol. 2016 Dec 5;238:153-164. doi: 10.1016/j.ijfoodmicro.2016.09.003. Epub 2016 Sep 8. Int J Food Microbiol. 2016. PMID: 27642688
-
Functional analysis of fungal polyketide biosynthesis genes.J Antibiot (Tokyo). 2010 May;63(5):207-18. doi: 10.1038/ja.2010.17. Epub 2010 Mar 5. J Antibiot (Tokyo). 2010. PMID: 20203700 Review.
-
Alternaria host-specific (HSTs) toxins: An overview of chemical characterization, target sites, regulation and their toxic effects.Toxicol Rep. 2019 Jul 17;6:745-758. doi: 10.1016/j.toxrep.2019.06.021. eCollection 2019. Toxicol Rep. 2019. PMID: 31406682 Free PMC article. Review.
Cited by
-
Comparative Genomics of Pathogens Causing Brown Spot Disease of Tobacco: Alternaria longipes and Alternaria alternata.PLoS One. 2016 May 9;11(5):e0155258. doi: 10.1371/journal.pone.0155258. eCollection 2016. PLoS One. 2016. PMID: 27159564 Free PMC article.
-
Draft Genome Sequence of Alternaria alternata Isolated from Onion Leaves in South Africa.Genome Announc. 2016 Sep 22;4(5):e01022-16. doi: 10.1128/genomeA.01022-16. Genome Announc. 2016. PMID: 27660793 Free PMC article.
-
Natural resorcylic lactones derived from alternariol.Beilstein J Org Chem. 2024 Aug 30;20:2171-2207. doi: 10.3762/bjoc.20.187. eCollection 2024. Beilstein J Org Chem. 2024. PMID: 39224229 Free PMC article. Review.
-
Secondary metabolites in fungus-plant interactions.Front Plant Sci. 2015 Aug 6;6:573. doi: 10.3389/fpls.2015.00573. eCollection 2015. Front Plant Sci. 2015. PMID: 26300892 Free PMC article. Review.
-
Linking a polyketide synthase gene cluster to 6-pentyl-alpha-pyrone, a Trichoderma metabolite with diverse bioactivities.Microb Cell Fact. 2025 Apr 21;24(1):89. doi: 10.1186/s12934-025-02718-9. Microb Cell Fact. 2025. PMID: 40259335 Free PMC article.
References
-
- Scott PM. Analysis of agricultural commodities and foods for Alternaria mycotoxins. J AOAC Int. 2001;84:1809–1817. - PubMed
-
- Ackermann Y, Curtui V, Dietrich R, Gross M, Lativ H, et al. Widespread occurrence of low levels of alternariol in apple and tomato products, as determined by comparative immunochemical assessment using monoclonal and polyclonal antibodies. J Agric Food Chem. 2011;59:6360–6368. - PubMed
-
- Liu GT, Quian YZ, Zhang P, Dong WH, Qi YM, et al. Etiological role of Alternaria alternata in human esophageal cancer. Chin Med J. 1992;105:394–400. - PubMed
-
- Brugger EM, Wagner J, Schumacher DM, Koch K, Podlech J, et al. Mutagenicity of the mycotoxin alternariol in cultured mammalian cells. Toxicol Lett. 2006;164:221–230. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

