A beta-herpesvirus with fluorescent capsids to study transport in living cells
- PMID: 22792376
- PMCID: PMC3394720
- DOI: 10.1371/journal.pone.0040585
A beta-herpesvirus with fluorescent capsids to study transport in living cells
Abstract
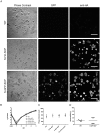
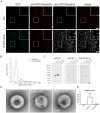
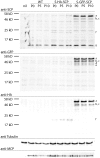
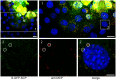
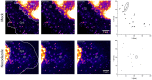
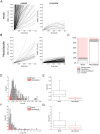
Fluorescent tagging of viral particles by genetic means enables the study of virus dynamics in living cells. However, the study of beta-herpesvirus entry and morphogenesis by this method is currently limited. This is due to the lack of replication competent, capsid-tagged fluorescent viruses. Here, we report on viable recombinant MCMVs carrying ectopic insertions of the small capsid protein (SCP) fused to fluorescent proteins (FPs). The FPs were inserted into an internal position which allowed the production of viable, fluorescently labeled cytomegaloviruses, which replicated with wild type kinetics in cell culture. Fluorescent particles were readily detectable by several methods. Moreover, in a spread assay, labeled capsids accumulated around the nucleus of the newly infected cells without any detectable viral gene expression suggesting normal entry and particle trafficking. These recombinants were used to record particle dynamics by live-cell microscopy during MCMV egress with high spatial as well as temporal resolution. From the resulting tracks we obtained not only mean track velocities but also their mean square displacements and diffusion coefficients. With this key information, we were able to describe particle behavior at high detail and discriminate between particle tracks exhibiting directed movement and tracks in which particles exhibited free or anomalous diffusion.
Conflict of interest statement
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

