Multiple sex-associated regions and a putative sex chromosome in zebrafish revealed by RAD mapping and population genomics
- PMID: 22792396
- PMCID: PMC3392230
- DOI: 10.1371/journal.pone.0040701
Multiple sex-associated regions and a putative sex chromosome in zebrafish revealed by RAD mapping and population genomics
Abstract
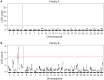
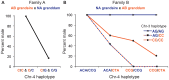
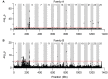
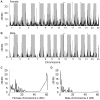
Within vertebrates, major sex determining genes can differ among taxa and even within species. In zebrafish (Danio rerio), neither heteromorphic sex chromosomes nor single sex determination genes of large effect, like Sry in mammals, have yet been identified. Furthermore, environmental factors can influence zebrafish sex determination. Although progress has been made in understanding zebrafish gonad differentiation (e.g. the influence of germ cells on gonad fate), the primary genetic basis of zebrafish sex determination remains poorly understood. To identify genetic loci associated with sex, we analyzed F(2) offspring of reciprocal crosses between Oregon *AB and Nadia (NA) wild-type zebrafish stocks. Genome-wide linkage analysis, using more than 5,000 sequence-based polymorphic restriction site associated (RAD-tag) markers and population genomic analysis of more than 30,000 single nucleotide polymorphisms in our *ABxNA crosses revealed a sex-associated locus on the end of the long arm of chr-4 for both cross families, and an additional locus in the middle of chr-3 in one cross family. Additional sequencing showed that two SNPs in dmrt1 previously suggested to be functional candidates for sex determination in a cross of ABxIndia wild-type zebrafish, are not associated with sex in our AB fish. Our data show that sex determination in zebrafish is polygenic and that different genes may influence sex determination in different strains or that different genes become more important under different environmental conditions. The association of the end of chr-4 with sex is remarkable because, unique in the karyotype, this chromosome arm shares features with known sex chromosomes: it is highly heterochromatic, repetitive, late replicating, and has reduced recombination. Our results reveal that chr-4 has functional and structural properties expected of a sex chromosome.
Conflict of interest statement
Figures

References
-
- Sinclair AH, Berta P, Palmer MS, Hawkins JR, Griffiths BL, et al. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature. 1990;346:240–244. - PubMed
-
- Bull JJ, Vogt RC. Temperature-dependent sex determination in turtles. Science. 1979;206:1186–1188. - PubMed
-
- Ferguson MW, Joanen T. Temperature of egg incubation determines sex in Alligator mississippiensis. Nature. 1982;296:850–853. - PubMed
-
- Crews D, Wibbels T, Gutzke WH. Action of sex steroid hormones on temperature-induced sex determination in the snapping turtle (Chelydra serpentina). Gen Comp Endocrinol. 1989;76:159–166. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

