Identification and functional studies of a new Nrf2 partner IQGAP1: a critical role in the stability and transactivation of Nrf2
- PMID: 22793650
- PMCID: PMC3689176
- DOI: 10.1089/ars.2012.4586
Identification and functional studies of a new Nrf2 partner IQGAP1: a critical role in the stability and transactivation of Nrf2
Abstract
Aims: Nuclear factor-erythroid-related factor 2 (Nrf2) is a critical transcriptional factor that is used in regulating cellular defense against oxidative stress. This study is aimed at investigating new interacting protein partners of Nrf2 using One-strep tag pull-down coupled with LTQ Orbitrap LC/MS/MS, and at examining the impact on Nr2 signaling by the newly identified IQ motif containing GTPase activating protein 1 (IQGAP1).
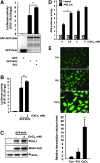
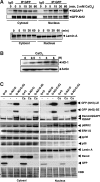
Results: Using the One-strep tag pull-down and LTQ Orbitrap LC/MS/MS, we identified IQGAP1 as a new Nrf2 interacting partner. Direct interactions between IQGAP1 and Nrf2 proteins were verified using in vitro glutathione S-transferase (GST) pull-down, transcription/translation assays, and in vivo utilizing Nrf2 overexpressing cells. Coexpression of Dsredmono-IQGAP1 and eGFP-Nrf2 increased the stability of eGFP-Nrf2 and enhanced the expression of Nrf2-target gene heme oxygenase-1 (HO-1). To confirm the functional role of IQGAP1 on Nrf2, knock-downed IQGAP1 using siIQGAP1 attenuated the expression of endogenous Nrf2, HO-1 proteins, and Nrf2-target genes GSTpi, GCLC, and
Nad(p)h: quinone oxidoreductase 1 (NQO-1). Furthermore, the stability of Nrf2 was dramatically decreased in IQGAP1-deficient mouse embryonic fibroblast (MEF) cells. Since IQGAP1 signaling could be mediated by calcium, treating the cells with calcium showed the translocation of IQGAP1/Nrf2 complex into the nucleus, suggesting that IQGAP1 may play a critical role in Nrf2 stability. Interestingly, consistent with calcium signaling for IQGAP1, treating the cells with calcium functionally enhanced Nrf2-mediated antioxidant responsive element-transcription activity and enhanced the expression of the endogenous Nrf2-target gene HO-1.
Innovation: In the aggregate, our current study identifies and functionally characterizes a new Nrf2 partner protein IQGAP1, which may contribute to Nrf2's regulation of antioxidant enzymes such as HO-1.
Conclusion: IQGAP1 may play a critical role in the stability and transactivation of Nrf2.
Figures



References
-
- Alam J. Stewart D. Touchard C. Boinapally S. Choi AM. Cook JL. Nrf2, a Cap'n'Collar transcription factor, regulates induction of the heme oxygenase-1 gene. J Biol Chem. 1999;274:26071–26078. - PubMed
-
- Berridge MJ. Lipp P. Bootman MD. The versatility and universality of calcium signalling. Nat Rev Mol Cell Biol. 2000;1:11–21. - PubMed
-
- Blank V. Small Maf proteins in mammalian gene control: mere dimerization partners or dynamic transcriptional regulators? J Mol Biol. 2008;376:913–925. - PubMed
-
- Briggs MW. Sacks DB. IQGAP1 as signal integrator: Ca2+, calmodulin, Cdc42 and the cytoskeleton. FEBS Lett. 2003;542:7–11. - PubMed
-
- Brown MD. Sacks DB. IQGAP1 in cellular signaling: bridging the GAP. Trends Cell Biol. 2006;16:242–249. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

