Multifunctionality and diversity of GDSL esterase/lipase gene family in rice (Oryza sativa L. japonica) genome: new insights from bioinformatics analysis
- PMID: 22793791
- PMCID: PMC3412167
- DOI: 10.1186/1471-2164-13-309
Multifunctionality and diversity of GDSL esterase/lipase gene family in rice (Oryza sativa L. japonica) genome: new insights from bioinformatics analysis
Abstract
Background: GDSL esterases/lipases are a newly discovered subclass of lipolytic enzymes that are very important and attractive research subjects because of their multifunctional properties, such as broad substrate specificity and regiospecificity. Compared with the current knowledge regarding these enzymes in bacteria, our understanding of the plant GDSL enzymes is very limited, although the GDSL gene family in plant species include numerous members in many fully sequenced plant genomes. Only two genes from a large rice GDSL esterase/lipase gene family were previously characterised, and the majority of the members remain unknown. In the present study, we describe the rice OsGELP (Oryza sativa GDSL esterase/lipase protein) gene family at the genomic and proteomic levels, and use this knowledge to provide insights into the multifunctionality of the rice OsGELP enzymes.
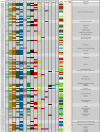
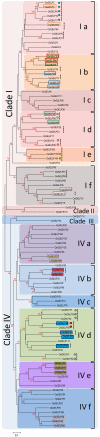
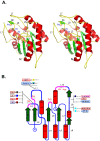
Results: In this study, an extensive bioinformatics analysis identified 114 genes in the rice OsGELP gene family. A complete overview of this family in rice is presented, including the chromosome locations, gene structures, phylogeny, and protein motifs. Among the OsGELPs and the plant GDSL esterase/lipase proteins of known functions, 41 motifs were found that represent the core secondary structure elements or appear specifically in different phylogenetic subclades. The specification and distribution of identified putative conserved clade-common and -specific peptide motifs, and their location on the predicted protein three dimensional structure may possibly signify their functional roles. Potentially important regions for substrate specificity are highlighted, in accordance with protein three-dimensional model and location of the phylogenetic specific conserved motifs. The differential expression of some representative genes were confirmed by quantitative real-time PCR. The phylogenetic analysis, together with protein motif architectures, and the expression profiling were analysed to predict the possible biological functions of the rice OsGELP genes.
Conclusions: Our current genomic analysis, for the first time, presents fundamental information on the organization of the rice OsGELP gene family. With combination of the genomic, phylogenetic, microarray expression, protein motif distribution, and protein structure analyses, we were able to create supported basis for the functional prediction of many members in the rice GDSL esterase/lipase family. The present study provides a platform for the selection of candidate genes for further detailed functional study.
Figures



Similar articles
-
Genome-wide analysis of GDSL-type esterases/lipases in Arabidopsis.Plant Mol Biol. 2017 Sep;95(1-2):181-197. doi: 10.1007/s11103-017-0648-y. Epub 2017 Aug 24. Plant Mol Biol. 2017. PMID: 28840447
-
Haplotype shifts in the lipid-related OsGELP gene family underpin rice adaptation to high latitudes.Sci Rep. 2025 Sep 2;15(1):32293. doi: 10.1038/s41598-025-15488-6. Sci Rep. 2025. PMID: 40897734 Free PMC article.
-
GDSL family of serine esterases/lipases.Prog Lipid Res. 2004 Nov;43(6):534-52. doi: 10.1016/j.plipres.2004.09.002. Prog Lipid Res. 2004. PMID: 15522763 Review.
-
Combining comparative sequence and genomic data to ascertain phylogenetic relationships and explore the evolution of the large GDSL-lipase family in land plants.Mol Biol Evol. 2011 Jan;28(1):551-65. doi: 10.1093/molbev/msq226. Epub 2010 Aug 26. Mol Biol Evol. 2011. PMID: 20801908
-
Genome-Wide Classification and Phylogenetic Analyses of the GDSL-Type Esterase/Lipase (GELP) Family in Flowering Plants.Int J Mol Sci. 2022 Oct 11;23(20):12114. doi: 10.3390/ijms232012114. Int J Mol Sci. 2022. PMID: 36292971 Free PMC article. Review.
Cited by
-
The hot pepper (Capsicum annuum) microRNA transcriptome reveals novel and conserved targets: a foundation for understanding MicroRNA functional roles in hot pepper.PLoS One. 2013 May 30;8(5):e64238. doi: 10.1371/journal.pone.0064238. Print 2013. PLoS One. 2013. PMID: 23737975 Free PMC article.
-
The DROOPING LEAF (DR) gene encoding GDSL esterase is involved in silica deposition in rice (Oryza sativa L.).PLoS One. 2020 Sep 10;15(9):e0238887. doi: 10.1371/journal.pone.0238887. eCollection 2020. PLoS One. 2020. PMID: 32913358 Free PMC article.
-
OsGELP77, a QTL for broad-spectrum disease resistance and yield in rice, encodes a GDSL-type lipase.Plant Biotechnol J. 2024 May;22(5):1352-1371. doi: 10.1111/pbi.14271. Epub 2023 Dec 15. Plant Biotechnol J. 2024. PMID: 38100249 Free PMC article.
-
Identification, evolution, and expression of GDSL-type Esterase/Lipase (GELP) gene family in three cotton species: a bioinformatic analysis.BMC Genomics. 2023 Dec 21;24(1):795. doi: 10.1186/s12864-023-09717-3. BMC Genomics. 2023. PMID: 38129780 Free PMC article.
-
Bioinformatics-Based Screening Approach for the Identification and Characterization of Lipolytic Enzymes from the Marine Diatom Phaeodactylum tricornutum.Mar Drugs. 2023 Feb 14;21(2):125. doi: 10.3390/md21020125. Mar Drugs. 2023. PMID: 36827166 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

