BiP-mediated closing of the Sec61 channel limits Ca2+ leakage from the ER
- PMID: 22796945
- PMCID: PMC3411083
- DOI: 10.1038/emboj.2012.189
BiP-mediated closing of the Sec61 channel limits Ca2+ leakage from the ER
Erratum in
- EMBO J. 2012 Sep 12;31(18):3784
Abstract
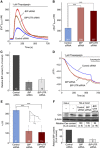
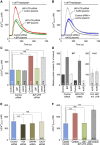
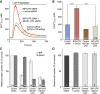
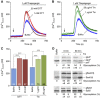
In mammalian cells, signal peptide-dependent protein transport into the endoplasmic reticulum (ER) is mediated by a dynamic protein-conducting channel, the Sec61 complex. Previous work has characterized the Sec61 channel as a potential ER Ca(2+) leak channel and identified calmodulin as limiting Ca(2+) leakage in a Ca(2+)-dependent manner by binding to an IQ motif in the cytosolic aminoterminus of Sec61α. Here, we manipulated the concentration of the ER lumenal chaperone BiP in cells in different ways and used live cell Ca(2+) imaging to monitor the effects of reduced levels of BiP on ER Ca(2+) leakage. Regardless of how the BiP concentration was lowered, the absence of available BiP led to increased Ca(2+) leakage via the Sec61 complex. When we replaced wild-type Sec61α with mutant Sec61αY344H in the same model cell, however, Ca(2+) leakage from the ER increased and was no longer affected by manipulation of the BiP concentration. Thus, BiP limits ER Ca(2+) leakage through the Sec61 complex by binding to the ER lumenal loop 7 of Sec61α in the vicinity of tyrosine 344.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

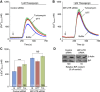
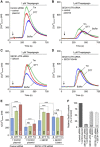

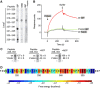

References
-
- Becker T, Bhushan S, Jarasch A, Armache JP, Funedad S, Jossinet F, Gumbart J, Mielke T, Berninghausen O, Schulten K, Westhof E, Gilmore R, Mandon ES, Beckmann R (2009) Structure of monomeric yeast and mammalian Sec61 complexes interacting with the translating ribosome. Science 326: 1369–1373 - PMC - PubMed
-
- Berridge MJ (2002) The endoplasmic reticulum: a multifunctional signalling organelle. Cell Calcium 32: 235–249 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

