The histone methyltransferase MMSET/WHSC1 activates TWIST1 to promote an epithelial-mesenchymal transition and invasive properties of prostate cancer
- PMID: 22797064
- PMCID: PMC3495247
- DOI: 10.1038/onc.2012.297
The histone methyltransferase MMSET/WHSC1 activates TWIST1 to promote an epithelial-mesenchymal transition and invasive properties of prostate cancer
Abstract
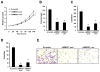
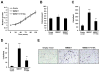
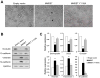
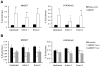
Epigenetic deregulation of gene expression has a role in the initiation and progression of prostate cancer (PCa). The histone methyltransferase MMSET/WHSC1 (Multiple Myeloma SET domain) is overexpressed in a number of metastatic tumors, but its mechanism of action has not been defined. In this work, we found that PCa cell lines expressed significantly higher levels of MMSET compared with immortalized, non-transformed prostate cells. Knockdown experiments showed that, in metastatic PCa cell lines, dimethylation of lysine 36 and trimethylation of lysine 27 on histone H3 (H3K36me2 and H3K27me3, respectively) depended on MMSET expression, whereas depletion of MMSET in benign prostatic cells did not affect chromatin modifications. Knockdown of MMSET in DU145 and PC-3 tumor cells decreased cell proliferation, colony formation in soft agar and strikingly diminished cell migration and invasion. Conversely, overexpression of MMSET in immortalized, non-transformed RWPE-1 cells promoted cell migration and invasion, accompanied by an epithelial-mesenchymal transition (EMT). Among a panel of EMT-promoting genes analyzed, TWIST1 expression was strongly activated in response to MMSET. Chromatin immunoprecipitation analysis demonstrated that MMSET binds to the TWIST1 locus and leads to an increase in H3K36me2, suggesting a direct role of MMSET in the regulation of this gene. Depletion of TWIST1 in MMSET-overexpressing RWPE-1 cells blocked cell invasion and EMT, indicating that TWIST1 was a critical target of MMSET, responsible for the acquisition of an invasive phenotype. Collectively, these data suggest that MMSET has a role in PCa pathogenesis and progression through epigenetic regulation of metastasis-related genes.
Conflict of interest statement
The authors declare no competing conflicts of interest.
Figures



Similar articles
-
Metformin Elicits Antitumor Effects and Downregulates the Histone Methyltransferase Multiple Myeloma SET Domain (MMSET) in Prostate Cancer Cells.Prostate. 2016 Dec;76(16):1507-1518. doi: 10.1002/pros.23235. Epub 2016 Jul 12. Prostate. 2016. PMID: 27404348
-
MiR-2392 suppresses metastasis and epithelial-mesenchymal transition by targeting MAML3 and WHSC1 in gastric cancer.FASEB J. 2017 Sep;31(9):3774-3786. doi: 10.1096/fj.201601140RR. Epub 2017 May 16. FASEB J. 2017. PMID: 28512191
-
MMSET/WHSC1 enhances DNA damage repair leading to an increase in resistance to chemotherapeutic agents.Oncogene. 2016 Nov 10;35(45):5905-5915. doi: 10.1038/onc.2016.116. Epub 2016 Apr 25. Oncogene. 2016. PMID: 27109101 Free PMC article.
-
NSD2 as a Promising Target in Hematological Disorders.Int J Mol Sci. 2022 Sep 21;23(19):11075. doi: 10.3390/ijms231911075. Int J Mol Sci. 2022. PMID: 36232375 Free PMC article. Review.
-
Normal and disease-related biological functions of Twist1 and underlying molecular mechanisms.Cell Res. 2012 Jan;22(1):90-106. doi: 10.1038/cr.2011.144. Epub 2011 Aug 30. Cell Res. 2012. PMID: 21876555 Free PMC article. Review.
Cited by
-
NSD2 is a requisite subunit of the AR/FOXA1 neo-enhanceosome in promoting prostate tumorigenesis.Nat Genet. 2024 Oct;56(10):2132-2143. doi: 10.1038/s41588-024-01893-6. Epub 2024 Sep 9. Nat Genet. 2024. PMID: 39251788 Free PMC article.
-
Relationship of Signaling Pathways between RKIP Expression and the Inhibition of EMT-Inducing Transcription Factors SNAIL1/2, TWIST1/2 and ZEB1/2.Cancers (Basel). 2024 Sep 17;16(18):3180. doi: 10.3390/cancers16183180. Cancers (Basel). 2024. PMID: 39335152 Free PMC article. Review.
-
Epidermal growth factor signaling in transformed cells.Int Rev Cell Mol Biol. 2015;314:1-41. doi: 10.1016/bs.ircmb.2014.10.001. Epub 2014 Nov 18. Int Rev Cell Mol Biol. 2015. PMID: 25619714 Free PMC article. Review.
-
NSD2 is a conserved driver of metastatic prostate cancer progression.Nat Commun. 2018 Dec 5;9(1):5201. doi: 10.1038/s41467-018-07511-4. Nat Commun. 2018. PMID: 30518758 Free PMC article.
-
Targeting epigenetic regulators for cancer therapy: mechanisms and advances in clinical trials.Signal Transduct Target Ther. 2019 Dec 17;4:62. doi: 10.1038/s41392-019-0095-0. eCollection 2019. Signal Transduct Target Ther. 2019. PMID: 31871779 Free PMC article. Review.
References
-
- Jemal A, Siegel R, Xu J, Ward E. Cancer statistics. CA Cancer J Clin. 2010;60:277–300. - PubMed
-
- Jeronimo C, Bastian PJ, Bjartell A, et al. Epigenetics in prostate cancer: biologic and clinical relevance. Eur Urol. 60:753–66. - PubMed
-
- Perry AS, Watson RW, Lawler M, Hollywood D. The epigenome as a therapeutic target in prostate cancer. Nat Rev Urol. 7:668–80. - PubMed
-
- Yegnasubramanian S, Kowalski J, Gonzalgo ML, et al. Hypermethylation of CpG islands in primary and metastatic human prostate cancer. Cancer Res. 2004;64:1975–86. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

