The Blk pathway functions as a tumor suppressor in chronic myeloid leukemia stem cells
- PMID: 22797726
- PMCID: PMC3408839
- DOI: 10.1038/ng.2350
The Blk pathway functions as a tumor suppressor in chronic myeloid leukemia stem cells
Abstract
A therapeutic strategy for treating cancer is to target and eradicate cancer stem cells (CSCs) without harming their normal stem cell counterparts. The success of this approach relies on the identification of molecular pathways that selectively regulate CSC function. Using BCR-ABL-induced chronic myeloid leukemia (CML) as a disease model for CSCs, we show that BCR-ABL downregulates the Blk gene (encoding B-lymphoid kinase) through c-Myc in leukemic stem cells (LSCs) in CML mice and that Blk functions as a tumor suppressor in LSCs but does not affect normal hematopoietic stem cells (HSCs) or hematopoiesis. Blk suppresses LSC function through a pathway involving an upstream regulator, Pax5, and a downstream effector, p27. Inhibition of this Blk pathway accelerates CML development, whereas increased activity of the Blk pathway delays CML development. Blk also suppresses the proliferation of human CML stem cells. Our results show the feasibility of selectively targeting LSCs, an approach that should be applicable to other cancers.
Conflict of interest statement
Authors have no competing financial interests.
Figures

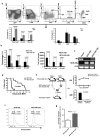


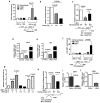

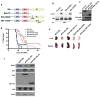

Similar articles
-
Dual targeting of p53 and c-MYC selectively eliminates leukaemic stem cells.Nature. 2016 Jun 16;534(7607):341-6. doi: 10.1038/nature18288. Epub 2016 Jun 8. Nature. 2016. PMID: 27281222 Free PMC article.
-
The Alox5 gene is a novel therapeutic target in cancer stem cells of chronic myeloid leukemia.Cell Cycle. 2009 Nov 1;8(21):3488-92. doi: 10.4161/cc.8.21.9852. Epub 2009 Nov 19. Cell Cycle. 2009. PMID: 19823023
-
PTEN is a tumor suppressor in CML stem cells and BCR-ABL-induced leukemias in mice.Blood. 2010 Jan 21;115(3):626-35. doi: 10.1182/blood-2009-06-228130. Epub 2009 Nov 18. Blood. 2010. PMID: 19965668 Free PMC article.
-
Molecular mechanisms for survival regulation of chronic myeloid leukemia stem cells.Protein Cell. 2013 Mar;4(3):186-96. doi: 10.1007/s13238-013-2115-0. Epub 2013 Mar 13. Protein Cell. 2013. PMID: 23483480 Free PMC article. Review.
-
Molecular pathogenesis of chronic myeloid leukemia: implications for new therapeutic strategies.Ann Hematol. 1999 Feb;78(2):49-64. doi: 10.1007/s002770050473. Ann Hematol. 1999. PMID: 10089019 Review.
Cited by
-
Emerging Kinase Therapeutic Targets in Pancreatic Ductal Adenocarcinoma and Pancreatic Cancer Desmoplasia.Int J Mol Sci. 2020 Nov 21;21(22):8823. doi: 10.3390/ijms21228823. Int J Mol Sci. 2020. PMID: 33233470 Free PMC article. Review.
-
Depletion of γ-catenin by Histone Deacetylase Inhibition Confers Elimination of CML Stem Cells in Combination with Imatinib.Theranostics. 2016 Aug 12;6(11):1947-62. doi: 10.7150/thno.16139. eCollection 2016. Theranostics. 2016. PMID: 27570562 Free PMC article.
-
The non-genomic loss of function of tumor suppressors: an essential role in the pathogenesis of chronic myeloid leukemia chronic phase.BMC Cancer. 2016 May 16;16:314. doi: 10.1186/s12885-016-2346-6. BMC Cancer. 2016. PMID: 27184141 Free PMC article. Review.
-
Tyrosine phosphorylation of IRF3 by BLK facilitates its sufficient activation and innate antiviral response.PLoS Pathog. 2023 Oct 23;19(10):e1011742. doi: 10.1371/journal.ppat.1011742. eCollection 2023 Oct. PLoS Pathog. 2023. PMID: 37871014 Free PMC article.
-
Cks1 is a critical regulator of hematopoietic stem cell quiescence and cycling, operating upstream of Cdk inhibitors.Oncogene. 2015 Aug 13;34(33):4347-57. doi: 10.1038/onc.2014.364. Epub 2014 Nov 24. Oncogene. 2015. PMID: 25417705
References
-
- Huntly BJ, Gilliland DG. Leukaemia stem cells and the evolution of cancer-stem-cell research. Nat Rev Cancer. 2005;5:311–21. - PubMed
-
- Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–11. - PubMed
-
- Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8:755–68. - PubMed
-
- Wang JC, Dick JE. Cancer stem cells: lessons from leukemia. Trends Cell Biol. 2005;15:494–501. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

