Peroxide-sensing transcriptional regulators in bacteria
- PMID: 22797754
- PMCID: PMC3458676
- DOI: 10.1128/JB.00304-12
Peroxide-sensing transcriptional regulators in bacteria
Abstract
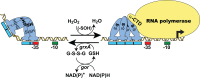
The ability to maintain intracellular concentrations of toxic reactive oxygen species (ROS) within safe limits is essential for all aerobic life forms. In bacteria, as well as other organisms, ROS are produced during the normal course of aerobic metabolism, necessitating the constitutive expression of ROS scavenging systems. However, bacteria can also experience transient high-level exposure to ROS derived either from external sources, such as the host defense response, or as a secondary effect of other seemingly unrelated environmental stresses. Consequently, transcriptional regulators have evolved to sense the levels of ROS and coordinate the appropriate oxidative stress response. Three well-studied examples of these are the peroxide responsive regulators OxyR, PerR, and OhrR. OxyR and PerR are sensors of primarily H(2)O(2), while OhrR senses organic peroxide (ROOH) and sodium hypochlorite (NaOCl). OxyR and OhrR sense oxidants by means of the reversible oxidation of specific cysteine residues. In contrast, PerR senses H(2)O(2) via the Fe-catalyzed oxidation of histidine residues. These transcription regulators also influence complex biological phenomena, such as biofilm formation, the evasion of host immune responses, and antibiotic resistance via the direct regulation of specific proteins.
Figures

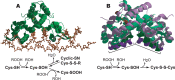
References
-
- Bae HW, Cho YH. 2012. Mutational analysis of Pseudomonas aeruginosa OxyR to define the regions required for peroxide resistance and acute virulence. Res. Microbiol. 163: 55–63 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

