Transcriptional regulation of the ecp operon by EcpR, IHF, and H-NS in attaching and effacing Escherichia coli
- PMID: 22797761
- PMCID: PMC3430341
- DOI: 10.1128/JB.00915-12
Transcriptional regulation of the ecp operon by EcpR, IHF, and H-NS in attaching and effacing Escherichia coli
Abstract
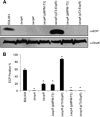
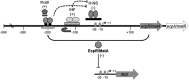
Enteropathogenic (EPEC) and enterohemorrhagic (EHEC) Escherichia coli are clinically important diarrheagenic pathogens that adhere to the intestinal epithelial surface. The E. coli common pili (ECP), or meningitis-associated and temperature-regulated (MAT) fimbriae, are ubiquitous among both commensal and pathogenic E. coli strains and play a role as colonization factors by promoting the interaction between bacteria and host epithelial cells and favoring interbacterial interactions in biofilm communities. The first gene of the ecp operon encodes EcpR (also known as MatA), a proposed regulatory protein containing a LuxR-like C-terminal helix-turn-helix (HTH) DNA-binding motif. In this work, we analyzed the transcriptional regulation of the ecp genes and the role of EcpR as a transcriptional regulator. EHEC and EPEC ecpR mutants produce less ECP, while plasmids expressing EcpR increase considerably the expression of EcpA and production of ECP. The ecp genes are transcribed as an operon from a promoter located 121 bp upstream of the start codon of ecpR. EcpR positively regulates this promoter by binding to two TTCCT boxes distantly located upstream of the ecp promoter, thus enhancing expression of downstream ecp genes, leading to ECP production. EcpR mutants in the putative HTH DNA-binding domain are no longer able to activate ecp expression or bind to the TTCCT boxes. EcpR-mediated activation is aided by integration host factor (IHF), which is essential for counteracting the repression exerted by histone-like nucleoid-structuring protein (H-NS) on the ecp promoter. This work demonstrates evidence about the interplay between a novel member of a diverse family of regulatory proteins and global regulators in the regulation of a fimbrial operon.
Figures






Similar articles
-
Finding Regulators Associated with the Expression of the Long Polar Fimbriae in Enteropathogenic Escherichia coli.J Bacteriol. 2015 Dec;197(23):3658-65. doi: 10.1128/JB.00509-15. Epub 2015 Sep 8. J Bacteriol. 2015. PMID: 26350135 Free PMC article.
-
Molecular characterization of GrlA, a specific positive regulator of ler expression in enteropathogenic Escherichia coli.J Bacteriol. 2010 Sep;192(18):4627-42. doi: 10.1128/JB.00307-10. Epub 2010 Jul 9. J Bacteriol. 2010. PMID: 20622062 Free PMC article.
-
Bacterial-Chromatin Structural Proteins Regulate the Bimodal Expression of the Locus of Enterocyte Effacement (LEE) Pathogenicity Island in Enteropathogenic Escherichia coli.mBio. 2017 Aug 8;8(4):e00773-17. doi: 10.1128/mBio.00773-17. mBio. 2017. PMID: 28790204 Free PMC article.
-
The Tip of the Iceberg: On the Roles of Regulatory Small RNAs in the Virulence of Enterohemorrhagic and Enteropathogenic Escherichia coli.Front Cell Infect Microbiol. 2016 Sep 21;6:105. doi: 10.3389/fcimb.2016.00105. eCollection 2016. Front Cell Infect Microbiol. 2016. PMID: 27709103 Free PMC article. Review.
-
Cytoskeleton-modulating effectors of enteropathogenic and enterohemorrhagic Escherichia coli: role of EspL2 in adherence and an alternative pathway for modulating cytoskeleton through Annexin A2 function.FEBS J. 2010 Jun;277(11):2403-8. doi: 10.1111/j.1742-4658.2010.07654.x. Epub 2010 Apr 30. FEBS J. 2010. PMID: 20477868 Review.
Cited by
-
Novel Avian Pathogenic Escherichia coli Genes Responsible for Adhesion to Chicken and Human Cell Lines.Appl Environ Microbiol. 2020 Oct 1;86(20):e01068-20. doi: 10.1128/AEM.01068-20. Print 2020 Oct 1. Appl Environ Microbiol. 2020. PMID: 32769194 Free PMC article.
-
The integration host factor regulates multiple virulence pathways in bacterial pathogen Dickeya zeae MS2.Mol Plant Pathol. 2022 Oct;23(10):1487-1507. doi: 10.1111/mpp.13244. Epub 2022 Jul 12. Mol Plant Pathol. 2022. PMID: 35819797 Free PMC article.
-
Exploitation of a "hockey-puck" phenotype to identify pilus and biofilm regulators in Serratia marcescens through genetic analysis.Can J Microbiol. 2016 Jan;62(1):83-93. doi: 10.1139/cjm-2015-0566. Epub 2015 Nov 3. Can J Microbiol. 2016. PMID: 26640000 Free PMC article.
-
Independent Component Analysis Identifies the Modulons Expanding the Transcriptional Regulatory Networks of Enterohemorrhagic Escherichia coli.Front Microbiol. 2022 Jun 24;13:953404. doi: 10.3389/fmicb.2022.953404. eCollection 2022. Front Microbiol. 2022. PMID: 35814713 Free PMC article.
-
The impact of Hfq-mediated sRNA-mRNA interactome on the virulence of enteropathogenic Escherichia coli.Sci Adv. 2021 Oct 29;7(44):eabi8228. doi: 10.1126/sciadv.abi8228. Epub 2021 Oct 27. Sci Adv. 2021. PMID: 34705501 Free PMC article.
References
-
- Aravind L, Anantharaman V, Balaji S, Babu MM, Iyer LM. 2005. The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol. Rev. 29:231–262 - PubMed
-
- Avelino F, et al. 2010. The majority of enteroaggregative Escherichia coli strains produce the E. coli common pilus when adhering to cultured epithelial cells. Int. J. Med. Microbiol. 300:440–448 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

