Neuroepithelial body microenvironment is a niche for a distinct subset of Clara-like precursors in the developing airways
- PMID: 22797898
- PMCID: PMC3412014
- DOI: 10.1073/pnas.1204710109
Neuroepithelial body microenvironment is a niche for a distinct subset of Clara-like precursors in the developing airways
Abstract
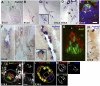
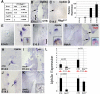
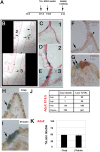
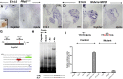
Clara cells of mammalian airways have multiple functions and are morphologically heterogeneous. Although Notch signaling is essential for the development of these cells, it is unclear how Notch influences Clara cell specification and if diversity is established among Clara cell precursors. Here we identify expression of the secretoglobin Scgb3a2 and Notch activation as early events in a program of secretory cell fate determination in developing murine airways. We show that Scgb3a2 expression in vivo is Notch-dependent at early stages and ectopically induced by constitutive Notch1 activation, and also that in vitro Notch signaling together with the pan-airway transcription factor Ttf1 (Nkx2.1) synergistically regulate secretoglobin gene transcription. Furthermore, we identified a subpopulation of secretory precursors juxtaposed to presumptive neuroepithelial bodies (NEBs), distinguished by their strong Scgb3a2 and uroplakin 3a (Upk3a) signals and reduced Ccsp (Scgb1a1) expression. Genetic ablation of Ascl1 prevented NEB formation and selectively interfered with the formation of this subpopulation of cells. Lineage labeling of Upk3a-expressing cells during development showed that these cells remain largely uncommitted during embryonic development and contribute to Clara and ciliated cells in the adult lung. Together, our findings suggest a role for Notch in the induction of a Clara cell-specific program of gene expression, and reveals that the NEB microenvironment in the developing airways is a niche for a distinct subset of Clara-like precursors.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Rawlins EL, et al. Epithelial stem/progenitor cells in lung postnatal growth, maintenance, and repair. Cold Spring Harb Symp Quant Biol. 2008;73:291–295. - PubMed
-
- Stripp BR, Maxson K, Mera R, Singh G. Plasticity of airway cell proliferation and gene expression after acute naphthalene injury. Am J Physiol. 1995;269:L791–L799. - PubMed
-
- Hung KS. Development of neuroepithelial bodies in pre- and postnatal mouse lungs: Scanning electron microscopic study. Anat Rec. 1982;203:285–291. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

