Molecular architecture and assembly principles of Vibrio cholerae biofilms
- PMID: 22798614
- PMCID: PMC3513368
- DOI: 10.1126/science.1222981
Molecular architecture and assembly principles of Vibrio cholerae biofilms
Abstract
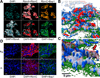
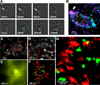
In their natural environment, microbes organize into communities held together by an extracellular matrix composed of polysaccharides and proteins. We developed an in vivo labeling strategy to allow the extracellular matrix of developing biofilms to be visualized with conventional and superresolution light microscopy. Vibrio cholerae biofilms displayed three distinct levels of spatial organization: cells, clusters of cells, and collections of clusters. Multiresolution imaging of living V. cholerae biofilms revealed the complementary architectural roles of the four essential matrix constituents: RbmA provided cell-cell adhesion; Bap1 allowed the developing biofilm to adhere to surfaces; and heterogeneous mixtures of Vibrio polysaccharide, RbmC, and Bap1 formed dynamic, flexible, and ordered envelopes that encased the cell clusters.
Figures

Comment in
-
Biofilms: Watching bacteria build their homes.Nat Rev Microbiol. 2012 Sep;10(9):597. doi: 10.1038/nrmicro2856. Epub 2012 Jul 30. Nat Rev Microbiol. 2012. PMID: 22842660 No abstract available.
References
-
- Costerton JW, Stewart PS, Greenberg EP. Bacterial Biofilms: A Common Cause of Persistent Infections. Science. 1999;284:1318–1322. - PubMed
-
- Hall-Stoodley L, Stoodley P. Biofilm formation and dispersal and the transmission of human pathogens. Trends in Microbiology. 2005;13:7–10. - PubMed
-
- Parsek MR, Singh PK. Bacterial biofilms: An Emerging Link to Disease Pathogenesis. Annu. Rev. Microbiol. 2003;57:677–701. - PubMed
-
- Stewart PS, William Costerton J. Antibiotic resistance of bacteria in biofilms. The Lancet. 2001;358:135–138. - PubMed
-
- Mah T-F, et al. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature. 2003;426:306–310. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

