Genetic diversity of expressed Plasmodium falciparum var genes from Tanzanian children with severe malaria
- PMID: 22799500
- PMCID: PMC3488018
- DOI: 10.1186/1475-2875-11-230
Genetic diversity of expressed Plasmodium falciparum var genes from Tanzanian children with severe malaria
Abstract
Background: Severe malaria has been attributed to the expression of a restricted subset of the var multi-gene family, which encodes for Plasmodium falciparum erythrocyte membrane protein 1 (PfEMP1). PfEMP1 mediates cytoadherence and sequestration of infected erythrocytes into the post-capillary venules of vital organs such as the brain, lung or placenta. var genes are highly diverse and can be classified in three major groups (ups A, B and C) and two intermediate groups (B/A and B/C) based on the genomic location, gene orientation and upstream sequences. The genetic diversity of expressed var genes in relation to severity of disease in Tanzanian children was analysed.
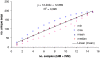
Methods: Children with defined severe (SM) and asymptomatic malaria (AM) were recruited. Full-length var mRNA was isolated and reversed transcribed into var cDNA. Subsequently, the DBL and N-terminal domains, and up-stream sequences were PCR amplified, cloned and sequenced. Sequences derived from SM and AM isolates were compared and analysed.
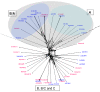
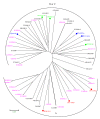
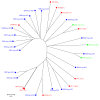
Results: The analysis confirmed that the var family is highly diverse in natural Plasmodium falciparum populations. Sequence diversity of amplified var DBL-1α and upstream regions showed minimal overlap among isolates, implying that the var gene repertoire is vast and most probably indefinite in endemic areas. var DBL-1α sequences from AM isolates were more diverse with more singletons found (p<0.05) than those from SM infections. Furthermore, few var DBL-1α sequences from SM patients were rare and restricted suggesting that certain PfEMP1 variants might induce severe disease.
Conclusions: The genetic sequence diversity of var genes of P. falciparum isolates from Tanzanian children is large and its relationship to disease severity has been studied. Observed differences suggest that different var genes might have fundamentally different roles in the host-parasite interaction. Further research is required to examine clear disease-associations of var gene subsets in different geographical settings. The importance of very strict clinical definitions and appropriate large control groups needs to be emphasized for future studies on disease associations of PfEMP1.
Figures
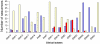

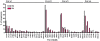
Similar articles
-
Diversity of the var gene family of Indonesian Plasmodium falciparum isolates.Malar J. 2013 Feb 27;12:80. doi: 10.1186/1475-2875-12-80. Malar J. 2013. PMID: 23446319 Free PMC article.
-
Meta-analysis of Plasmodium falciparum var Signatures Contributing to Severe Malaria in African Children and Indian Adults.mBio. 2019 Apr 30;10(2):e00217-19. doi: 10.1128/mBio.00217-19. mBio. 2019. PMID: 31040236 Free PMC article.
-
Sympatric Plasmodium falciparum isolates from Venezuela have structured var gene repertoires.Malar J. 2003 Apr 11;2:7. doi: 10.1186/1475-2875-2-7. Epub 2003 Apr 11. Malar J. 2003. PMID: 12737636 Free PMC article.
-
A systematic review on genetic diversity of var gene DBL1α domain from different geographical regions in Plasmodium falciparum isolates.Infect Genet Evol. 2021 Nov;95:105049. doi: 10.1016/j.meegid.2021.105049. Epub 2021 Aug 24. Infect Genet Evol. 2021. PMID: 34450294
-
var genes, PfEMP1 and the human host.Mol Biochem Parasitol. 2004 Mar;134(1):3-9. doi: 10.1016/j.molbiopara.2003.09.010. Mol Biochem Parasitol. 2004. PMID: 14747137 Review.
Cited by
-
In silico guided reconstruction and analysis of ICAM-1-binding var genes from Plasmodium falciparum.Sci Rep. 2018 Feb 19;8(1):3282. doi: 10.1038/s41598-018-21591-8. Sci Rep. 2018. PMID: 29459671 Free PMC article.
-
Evolutionary structure of Plasmodium falciparum major variant surface antigen genes in South America: Implications for epidemic transmission and surveillance.Ecol Evol. 2017 Oct 8;7(22):9376-9390. doi: 10.1002/ece3.3425. eCollection 2017 Nov. Ecol Evol. 2017. PMID: 29187975 Free PMC article.
-
Plasmodium falciparum expresses fewer var genes at lower levels during asymptomatic dry season infections than clinical malaria cases.PLoS Pathog. 2025 Jun 10;21(6):e1013210. doi: 10.1371/journal.ppat.1013210. eCollection 2025 Jun. PLoS Pathog. 2025. PMID: 40493675 Free PMC article.
-
CD4+ T cell responses to the Plasmodium falciparum erythrocyte membrane protein 1 in children with mild malaria.J Immunol. 2014 Feb 15;192(4):1753-61. doi: 10.4049/jimmunol.1200547. Epub 2014 Jan 22. J Immunol. 2014. PMID: 24453249 Free PMC article.
-
Genotyping var Gene DBL1α Domain of Severe and Non-severe Plasmodium falciparum Patients.Indian J Microbiol. 2024 Jun;64(2):583-592. doi: 10.1007/s12088-024-01200-1. Epub 2024 Feb 6. Indian J Microbiol. 2024. PMID: 39011004 Free PMC article.
References
-
- WHO. World malaria report: 2011. Geneva: World Health Organization; 2011.
-
- Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, Carlton JM, Pain A, Nelson KE, Bowman S, Paulsen IT, James K, Eisen JA, Rutherford K, Salzberg SL, Craig A, Kyes S, Chan MS, Nene V, Shallom SJ, Suh B, Peterson J, Angiuoli S, Pertea M, Allen J, Selengut J, Haft D, Mather MW, Vaidya AB, Martin DM, Fairlamb AH, Fraunholz MJ, Roos DS, Ralph SA, McFadden GI, Cummings LM, Subramanian GM, Mungall C, Venter JC, Carucci DJ, Hoffman SL, Newbold C, Davis RW, Fraser CM, Barrell B. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

