Transposable elements in phytopathogenic Verticillium spp.: insights into genome evolution and inter- and intra-specific diversification
- PMID: 22800085
- PMCID: PMC3441728
- DOI: 10.1186/1471-2164-13-314
Transposable elements in phytopathogenic Verticillium spp.: insights into genome evolution and inter- and intra-specific diversification
Abstract
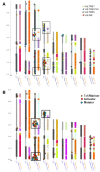
Background: Verticillium dahliae (Vd) and Verticillium albo-atrum (Va) are cosmopolitan soil fungi causing very disruptive vascular diseases on a wide range of crop plants. To date, no sexual stage has been identified in either microorganism suggesting that somatic mutation is a major force in generating genetic diversity. Whole genome comparative analysis of the recently sequenced strains VdLs.17 and VaMs.102 revealed that non-random insertions of transposable elements (TEs) have contributed to the generation of four lineage-specific (LS) regions in VdLs.17.
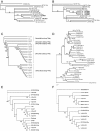
Results: We present here a detailed analysis of Class I retrotransposons and Class II "cut-and-paste" DNA elements detected in the sequenced Verticillium genomes. We report also of their distribution in other Vd and Va isolates from various geographic origins. In VdLs.17, we identified and characterized 56 complete retrotransposons of the Gypsy-, Copia- and LINE-like types, as well as 34 full-length elements of the "cut-and-paste" superfamilies Tc1/mariner, Activator and Mutator. While Copia and Tc1/mariner were present in multiple identical copies, Activator and Mutator sequences were highly divergent. Most elements comprised complete ORFs, had matching ESTs and showed active transcription in response to stress treatment. Noticeably, we found evidences of repeat-induced point mutation (RIP) only in some of the Gypsy retroelements. While Copia-, Gypsy- and Tc1/mariner-like transposons were prominent, a large variation in presence of the other types of mobile elements was detected in the other Verticillium spp. strains surveyed. In particular, neither complete nor defective "cut-and-paste" TEs were found in VaMs.102.
Conclusions: Copia-, Gypsy- and Tc1/mariner-like transposons are the most wide-spread TEs in the phytopathogens V. dahliae and V. albo-atrum. In VdLs.17, we identified several retroelements and "cut-and-paste" transposons still potentially active. Some of these elements have undergone diversification and subsequent selective amplification after introgression into the fungal genome. Others, such as the ripped Copias, have been potentially acquired by horizontal transfer. The observed biased TE insertion in gene-rich regions within an individual genome (VdLs.17) and the "patchy" distribution among different strains point to the mobile elements as major generators of Verticillium intra- and inter-specific genomic variation.
Figures


References
-
- Pegg GF, Brady BL. Verticillium wilts. CABI Publishing, New York; 2002.
-
- Klosterman SJ, Subbarao KV, Kang S, Veronese P, Gold SE, Thomma BPHJ, Chen Z, Henrissat B, Lee Y-H, Park J, Garcia-Pedrajas MD, Barbara DJ, Anchieta A, de Jonge R, Santhanam P, Maruthachalam K, Atallah Z, Amyotte SG, Paz Z, Inderbitzin P, Hayes RJ, Heiman DI, Young S, Zeng Q, Engels R, Galagan J, Cuomo CA, Dobinson KF, Ma L-J. Comparative genomics yields insights into niche adaptation of plant vascular wilt pathogens. PLoS Pathogens. 2011;7:e1002137. doi: 10.1371/journal.ppat.1002137. - DOI - PMC - PubMed
-
- Dobinson KF, Patterson NA, White GJ, Grant S. DNA fingerprinting and vegetative compatibility analysis indicate multiple origins for Verticillium dahliae race 2 tomato isolates from Ontario, Canada. Mycol Res. 1998;102:1089–1095. doi: 10.1017/S0953756297006035. - DOI
-
- Usami T, Shishido M, Ebihara Y, Kamigahira Y, Amemiya Y. Retrotransposon-like elements in the genome of Verticillium dahliae may be used as DNA markers for fungal species and pathotypes. J Gen Plant Pathol. 2005;71:117–123. doi: 10.1007/s10327-004-0171-2. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

