Paired-end sequencing of Fosmid libraries by Illumina
- PMID: 22800726
- PMCID: PMC3483553
- DOI: 10.1101/gr.138925.112
Paired-end sequencing of Fosmid libraries by Illumina
Abstract
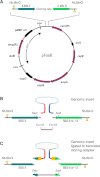
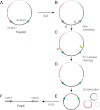
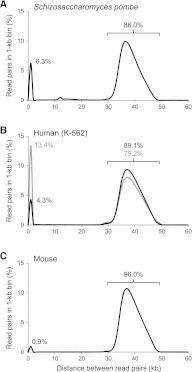
Eliminating the bacterial cloning step has been a major factor in the vastly improved efficiency of massively parallel sequencing approaches. However, this also has made it a technical challenge to produce the modern equivalent of the Fosmid- or BAC-end sequences that were crucial for assembling and analyzing complex genomes during the Sanger-based sequencing era. To close this technology gap, we developed Fosill, a method for converting Fosmids to Illumina-compatible jumping libraries. We constructed Fosmid libraries in vectors with Illumina primer sequences and specific nicking sites flanking the cloning site. Our family of pFosill vectors allows multiplex Fosmid cloning of end-tagged genomic fragments without physical size selection and is compatible with standard and multiplex paired-end Illumina sequencing. To excise the bulk of each cloned insert, we introduced two nicks in the vector, translated them into the inserts, and cleaved them. Recircularization of the vector via coligation of insert termini followed by inverse PCR generates a jumping library for paired-end sequencing with 101-base reads. The yield of unique Fosmid-sized jumps is sufficiently high, and the background of short, incorrectly spaced and chimeric artifacts sufficiently low, to enable applications such as mapping of structural variation and scaffolding of de novo assemblies. We demonstrate the power of Fosill to map genome rearrangements in a cancer cell line and identified three fusion genes that were corroborated by RNA-seq data. Our Fosill-powered assembly of the mouse genome has an N50 scaffold length of 17.0 Mb, rivaling the connectivity (16.9 Mb) of the Sanger-sequencing based draft assembly.
Figures
References
-
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al. 2000. The genome sequence of Drosophila melanogaster. Science 287: 2185–2195 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
