Hydrolase controls cellular NAD, sirtuin, and secondary metabolites
- PMID: 22801369
- PMCID: PMC3430197
- DOI: 10.1128/MCB.00032-12
Hydrolase controls cellular NAD, sirtuin, and secondary metabolites
Abstract
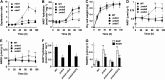
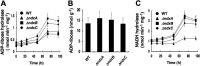
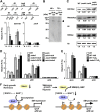
Cellular levels of NAD(+) and NADH are thought to be controlled by de novo and salvage mechanisms, although evidence has not yet indicated that they are regulated by NAD(+) degradation. Here we show that the conserved nudix hydrolase isozyme NdxA hydrolyzes and decreases cellular NAD(+) and NADH in Aspergillus nidulans. The NdxA-deficient fungus accumulated more NAD(+) during the stationary growth phase, indicating that NdxA maintains cellular NAD(+)/NADH homeostasis. The deficient strain also generated less of the secondary metabolites sterigmatocystin and penicillin G and of their gene transcripts than did the wild type. These defects were associated with a reduction in acetylated histone H4 on the gene promoters of aflR and ipnA that are involved in synthesizing secondary metabolites. Thus, NdxA increases acetylation levels of histone H4. We discovered that the novel fungal sirtuin isozyme SirA uses NAD(+) as a cosubstrate to deacetylate the lysine 16 residue of histone H4 on the gene promoter and represses gene expression. The impaired acetylation of histone and secondary metabolite synthesis in the NdxA-deficient strain were restored by eliminating functional SirA, indicating that SirA mediates NdxA-dependent regulation. These results indicated that NdxA controls total levels of NAD(+)/NADH and negatively regulates sirtuin function and chromatin structure.
Figures






Similar articles
-
Sirtuin E is a fungal global transcriptional regulator that determines the transition from the primary growth to the stationary phase.J Biol Chem. 2017 Jun 30;292(26):11043-11054. doi: 10.1074/jbc.M116.753772. Epub 2017 May 2. J Biol Chem. 2017. PMID: 28465348 Free PMC article.
-
Nudix hydrolase controls nucleotides and glycolytic mechanisms in hypoxic Aspergillus nidulans.Biosci Biotechnol Biochem. 2013;77(9):1888-93. doi: 10.1271/bbb.130334. Epub 2013 Sep 7. Biosci Biotechnol Biochem. 2013. PMID: 24018665
-
Sirtuin A regulates secondary metabolite production by Aspergillus nidulans.J Gen Appl Microbiol. 2017 Sep 5;63(4):228-235. doi: 10.2323/jgam.2016.11.002. Epub 2017 Jul 1. J Gen Appl Microbiol. 2017. PMID: 28674377
-
NAD+/NADH homeostasis affects metabolic adaptation to hypoxia and secondary metabolite production in filamentous fungi.Biosci Biotechnol Biochem. 2018 Feb;82(2):216-224. doi: 10.1080/09168451.2017.1422972. Epub 2018 Jan 12. Biosci Biotechnol Biochem. 2018. PMID: 29327656 Review.
-
Sirtuins in Epigenetic Silencing and Control of Gene Expression in Model and Pathogenic Fungi.Annu Rev Microbiol. 2022 Sep 8;76:157-178. doi: 10.1146/annurev-micro-041020-100926. Epub 2022 May 24. Annu Rev Microbiol. 2022. PMID: 35609947 Review.
Cited by
-
Fungus-specific sirtuin HstD coordinates secondary metabolism and development through control of LaeA.Eukaryot Cell. 2013 Aug;12(8):1087-96. doi: 10.1128/EC.00003-13. Epub 2013 May 31. Eukaryot Cell. 2013. PMID: 23729383 Free PMC article.
-
Regulation of Histone Acetylation Modification on Biosynthesis of Secondary Metabolites in Fungi.Int J Mol Sci. 2024 Dec 24;26(1):25. doi: 10.3390/ijms26010025. Int J Mol Sci. 2024. PMID: 39795886 Free PMC article. Review.
-
Sirtuin E is a fungal global transcriptional regulator that determines the transition from the primary growth to the stationary phase.J Biol Chem. 2017 Jun 30;292(26):11043-11054. doi: 10.1074/jbc.M116.753772. Epub 2017 May 2. J Biol Chem. 2017. PMID: 28465348 Free PMC article.
-
The human NAD metabolome: Functions, metabolism and compartmentalization.Crit Rev Biochem Mol Biol. 2015;50(4):284-97. doi: 10.3109/10409238.2015.1028612. Epub 2015 Apr 2. Crit Rev Biochem Mol Biol. 2015. PMID: 25837229 Free PMC article. Review.
-
Potential antifungal targets based on histones post-translational modifications against invasive aspergillosis.Front Microbiol. 2022 Aug 9;13:980615. doi: 10.3389/fmicb.2022.980615. eCollection 2022. Front Microbiol. 2022. PMID: 36016791 Free PMC article. Review.
References
-
- AbdelRaheim SR, Cartwright JL, Gasmi L, McLennan AG. 2001. The NADH diphosphatase encoded by the Saccharomyces cerevisiae NPY1 nudix hydrolase gene is located in peroxisomes. Arch. Biochem. Biophys. 388: 18–24 - PubMed
-
- Bayram O, et al. 2008. VelB/VeA/LaeA complex coordinates light signal with fungal development and secondary metabolism. Science 320: 1504–1506 - PubMed
-
- Belenky P, et al. 2007. Nicotinamide riboside promotes Sir2 silencing and extends lifespan via Nrk and Urh1/Pnp1/Meu1 pathways to NAD+. Cell 129: 473–484 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
