Existence of separate domains in lysin PlyG for recognizing Bacillus anthracis spores and vegetative cells
- PMID: 22802245
- PMCID: PMC3457386
- DOI: 10.1128/AAC.00891-12
Existence of separate domains in lysin PlyG for recognizing Bacillus anthracis spores and vegetative cells
Abstract
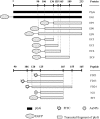
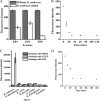
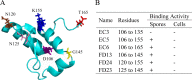
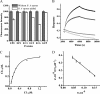
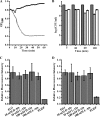
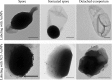
As a potential antimicrobial, the bacteriophage lysin PlyG has been reported to specifically recognize Bacillus anthracis vegetative cells only and to kill B. anthracis vegetative cells and its germinating spores. However, how PlyG interacts with B. anthracis spores remains unclear. Herein, a 60-amino-acid domain in PlyG (residues 106 to 165), located mainly in the previously identified catalytic domain, was found able to specifically recognize B. anthracis spores but not vegetative cells. The exosporium of the spores was found to be the most probable binding target of this domain. This is the first time that a lysin for spore-forming bacteria has been found to have separate domains to recognize spores and vegetative cells, which might help in understanding the coevolution of phages with spore-forming bacteria. Besides providing new biomarkers for developing better assays for identifying B. anthracis spores, the newly found domain may be helpful in developing PlyG as a preventive antibiotic to reduce the threat of anthrax in suspected exposures to B. anthracis spores.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

