Characterization of a rifampin-inactivating glycosyltransferase from a screen of environmental actinomycetes
- PMID: 22802246
- PMCID: PMC3457401
- DOI: 10.1128/AAC.01166-12
Characterization of a rifampin-inactivating glycosyltransferase from a screen of environmental actinomycetes
Abstract
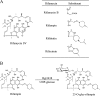
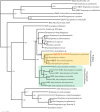
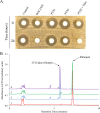
Identifying and understanding the collection of all antibiotic resistance determinants presented in the global microbiota, the antibiotic resistome, provides insight into the evolution of antibiotic resistance and critical information for the development of future antimicrobials. The rifamycins are broad-spectrum antibiotics that target bacterial transcription by inhibition of RNA polymerase. Although mutational alteration of the drug target is the predominant mechanism of resistance to this family of antibiotics in the clinic, a number of diverse inactivation mechanisms have also been reported. In this report, we investigate a subset of environmental rifampin-resistant actinomycete isolates and identify a diverse collection of rifampin inactivation mechanisms. We describe a single isolate, WAC1438, capable of inactivating rifampin by glycosylation. A draft genome sequence of WAC1438 (most closely related to Streptomyces speibonae, according to a 16S rRNA gene comparison) was assembled, and the associated rifampin glycosyltransferase open reading frame, rgt1438, was identified. The role of rgt1438 in rifampin resistance was confirmed by its disruption in the bacterial chromosome, resulting in a loss of antibiotic inactivation and a 4-fold decrease in MIC. Interestingly, examination of the RNA polymerase β-subunit sequence of WAC1438 suggests that it harbors a resistant target and thus possesses dual mechanisms of rifamycin resistance. Using an in vitro assay with purified enzyme, Rgt1438 could inactivate a variety of rifamycin antibiotics with comparable steady-state kinetics constants. Our results identify rgt1438 as a rifampin resistance determinant from WAC1438 capable of inactivating an assortment of rifamycins, adding a new element to the rifampin resistome.
Figures

Similar articles
-
HelR is a helicase-like protein that protects RNA polymerase from rifamycin antibiotics.Mol Cell. 2022 Sep 1;82(17):3151-3165.e9. doi: 10.1016/j.molcel.2022.06.019. Epub 2022 Jul 30. Mol Cell. 2022. PMID: 35907401
-
A rifamycin inactivating phosphotransferase family shared by environmental and pathogenic bacteria.Proc Natl Acad Sci U S A. 2014 May 13;111(19):7102-7. doi: 10.1073/pnas.1402358111. Epub 2014 Apr 28. Proc Natl Acad Sci U S A. 2014. PMID: 24778229 Free PMC article.
-
Rox, a Rifamycin Resistance Enzyme with an Unprecedented Mechanism of Action.Cell Chem Biol. 2018 Apr 19;25(4):403-412.e5. doi: 10.1016/j.chembiol.2018.01.009. Epub 2018 Feb 1. Cell Chem Biol. 2018. PMID: 29398560
-
The Enzymes of the Rifamycin Antibiotic Resistome.Acc Chem Res. 2021 May 4;54(9):2065-2075. doi: 10.1021/acs.accounts.1c00048. Epub 2021 Apr 20. Acc Chem Res. 2021. PMID: 33877820 Review.
-
Engineering the glycosylation of natural products in actinomycetes.Trends Microbiol. 2007 May;15(5):219-32. doi: 10.1016/j.tim.2007.03.004. Epub 2007 Apr 6. Trends Microbiol. 2007. PMID: 17412593 Review.
Cited by
-
Antibiotic resistome from the One-Health perspective: understanding and controlling antimicrobial resistance transmission.Exp Mol Med. 2021 Mar;53(3):301-309. doi: 10.1038/s12276-021-00569-z. Epub 2021 Mar 1. Exp Mol Med. 2021. PMID: 33642573 Free PMC article. Review.
-
Mechanism of Rifampicin Inactivation in Nocardia farcinica.PLoS One. 2016 Oct 5;11(10):e0162578. doi: 10.1371/journal.pone.0162578. eCollection 2016. PLoS One. 2016. PMID: 27706151 Free PMC article.
-
Antibacterial activity of and resistance to small molecule inhibitors of the ClpP peptidase.ACS Chem Biol. 2013 Dec 20;8(12):2669-77. doi: 10.1021/cb400577b. Epub 2013 Oct 4. ACS Chem Biol. 2013. PMID: 24047344 Free PMC article.
-
Arr-cb Is a Rifampin Resistance Determinant Found Active or Cryptic in Clostridium bolteae Strains.Antimicrob Agents Chemother. 2017 Jul 25;61(8):e00301-17. doi: 10.1128/AAC.00301-17. Print 2017 Aug. Antimicrob Agents Chemother. 2017. PMID: 28533241 Free PMC article.
-
Characterization of the soil resistome and mobilome in Namib Desert soils.Int Microbiol. 2024 Aug;27(4):967-975. doi: 10.1007/s10123-023-00454-x. Epub 2023 Nov 16. Int Microbiol. 2024. PMID: 37968548 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

