Multiple mutations in hepatitis C virus NS5A domain II are required to confer a significant level of resistance to alisporivir
- PMID: 22802259
- PMCID: PMC3457393
- DOI: 10.1128/AAC.00919-12
Multiple mutations in hepatitis C virus NS5A domain II are required to confer a significant level of resistance to alisporivir
Abstract
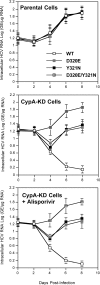
Alisporivir is the most advanced host-targeting antiviral cyclophilin (Cyp) inhibitor in phase III studies and has demonstrated a great deal of promise in decreasing hepatitis C virus (HCV) viremia in infected patients. In an attempt to further elucidate the mechanism of action of alisporivir, HCV replicons resistant to the drug were selected. Interestingly, mutations constantly arose in domain II of NS5A. To demonstrate that these mutations are responsible for drug resistance, they were reintroduced into the parental HCV genome, and the resulting mutant viruses were tested for replication in the presence of alisporivir or in the absence of the alisporivir target, CypA. We also examined the effect of the mutations on NS5A binding to itself (oligomerization), CypA, RNA, and NS5B. Importantly, the mutations did not affect any of these interactions. Moreover, the mutations did not preserve NS5A-CypA interactions from alisporivir rupture. NS5A mutations alone render HCV only slightly resistant to alisporivir. In sharp contrast, when multiple NS5A mutations are combined, significant resistance was observed. The introduction of multiple mutations in NS5A significantly restored viral replication in CypA knockdown cells. Interestingly, the combination of NS5A mutations renders HCV resistant to all classes of Cyp inhibitors. This study suggests that a combination of multiple mutations in domain II of NS5A rather than a single mutation is required to render HCV significantly and universally resistant to Cyp inhibitors. This in accordance with in vivo data that suggest that alisporivir is associated with a low potential for development of viral resistance.
Figures

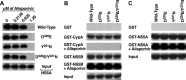

References
-
- Ahn J, et al. 2004. Systematic identification of hepatocellular proteins interacting with NS5A of the hepatitis C virus. J. Biochem. Mol. Biol. 37:741–748 - PubMed
-
- Armstrong GL, et al. 2006. The prevalence of hepatitis C virus infection in the United States, 1999 through 2002. Ann. Intern. Med. 144:705–714 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

