The atypical E2F family member E2F7 couples the p53 and RB pathways during cellular senescence
- PMID: 22802529
- PMCID: PMC3404383
- DOI: 10.1101/gad.196238.112
The atypical E2F family member E2F7 couples the p53 and RB pathways during cellular senescence
Abstract
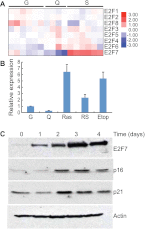
Oncogene-induced senescence is an anti-proliferative stress response program that acts as a fail-safe mechanism to limit oncogenic transformation and is regulated by the retinoblastoma protein (RB) and p53 tumor suppressor pathways. We identify the atypical E2F family member E2F7 as the only E2F transcription factor potently up-regulated during oncogene-induced senescence, a setting where it acts in response to p53 as a direct transcriptional target. Once induced, E2F7 binds and represses a series of E2F target genes and cooperates with RB to efficiently promote cell cycle arrest and limit oncogenic transformation. Disruption of RB triggers a further increase in E2F7, which induces a second cell cycle checkpoint that prevents unconstrained cell division despite aberrant DNA replication. Mechanistically, E2F7 compensates for the loss of RB in repressing mitotic E2F target genes. Together, our results identify a causal role for E2F7 in cellular senescence and uncover a novel link between the RB and p53 pathways.
Figures



References
-
- Attwooll C, Oddi S, Cartwright P, Prosperini E, Agger K, Steensgaard P, Wagener C, Sardet C, Moroni MC, Helin K 2005. A novel repressive E2F6 complex containing the polycomb group protein, EPC1, that interacts with EZH2 in a proliferation-specific manner. J Biol Chem 280: 1199–1208 - PubMed
-
- Azzoli CG, Sagar M, Wu A, Lowry D, Hennings H, Morgan DL, Weinberg WC 1998. Cooperation of p53 loss of function and v-Ha-ras in transformation of mouse keratinocyte cell lines. Mol Carcinog 21: 50–61 - PubMed
-
- Braig M, Lee S, Loddenkemper C, Rudolph C, Peters AH, Schlegelberger B, Stein H, Dörken B, Jenuwein T, Schmitt CA 2005. Oncogene-induced senescence as an initial barrier in lymphoma development. Nature 436: 660–665 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
