Structural basis for intersubunit signaling in a protein disaggregating machine
- PMID: 22802670
- PMCID: PMC3411974
- DOI: 10.1073/pnas.1207040109
Structural basis for intersubunit signaling in a protein disaggregating machine
Abstract
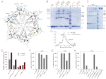
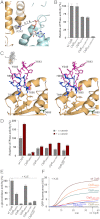
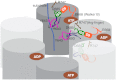
ClpB is a ring-forming, ATP-dependent protein disaggregase that cooperates with the cognate Hsp70 system to recover functional protein from aggregates. How ClpB harnesses the energy of ATP binding and hydrolysis to facilitate the mechanical unfolding of previously aggregated, stress-damaged proteins remains unclear. Here, we present crystal structures of the ClpB D2 domain in the nucleotide-bound and -free states, and the fitted cryoEM structure of the D2 hexamer ring, which provide a structural understanding of the ATP power stroke that drives protein translocation through the ClpB hexamer. We demonstrate that the conformation of the substrate-translocating pore loop is coupled to the nucleotide state of the cis subunit, which is transmitted to the neighboring subunit via a conserved but structurally distinct intersubunit-signaling pathway common to diverse AAA+ machines. Furthermore, we found that an engineered, disulfide cross-linked ClpB hexamer is fully functional biochemically, suggesting that ClpB deoligomerization is not required for protein disaggregation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

