The dengue vector Aedes aegypti contains a functional high mobility group box 1 (HMGB1) protein with a unique regulatory C-terminus
- PMID: 22802955
- PMCID: PMC3388995
- DOI: 10.1371/journal.pone.0040192
The dengue vector Aedes aegypti contains a functional high mobility group box 1 (HMGB1) protein with a unique regulatory C-terminus
Abstract
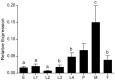
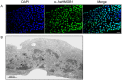
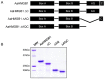
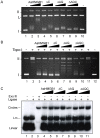
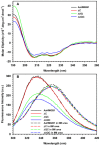
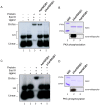
The mosquito Aedes aegypti can spread the dengue, chikungunya and yellow fever viruses. Thus, the search for key molecules involved in the mosquito survival represents today a promising vector control strategy. High Mobility Group Box (HMGB) proteins are essential nuclear factors that maintain the high-order structure of chromatin, keeping eukaryotic cells viable. Outside the nucleus, secreted HMGB proteins could alert the innate immune system to foreign antigens and trigger the initiation of host defenses. In this work, we cloned and functionally characterized the HMGB1 protein from Aedes aegypti (AaHMGB1). The AaHMGB1 protein typically consists of two HMG-box DNA binding domains and an acidic C-terminus. Interestingly, AaHMGB1 contains a unique alanine/glutamine-rich (AQ-rich) C-terminal region that seems to be exclusive of dipteran HMGB proteins. AaHMGB1 is localized to the cell nucleus, mainly associated with heterochromatin. Circular dichroism analyses of AaHMGB1 or the C-terminal truncated proteins revealed α-helical structures. We showed that AaHMGB1 can effectively bind and change the topology of DNA, and that the AQ-rich and the C-terminal acidic regions can modulate its ability to promote DNA supercoiling, as well as its preference to bind supercoiled DNA. AaHMGB1 is phosphorylated by PKA and PKC, but not by CK2. Importantly, phosphorylation of AaHMGB1 by PKA or PKC completely abolishes its DNA bending activity. Thus, our study shows that a functional HMGB1 protein occurs in Aedes aegypt and we provide the first description of a HMGB1 protein containing an AQ-rich regulatory C-terminus.
Conflict of interest statement
Figures


References
-
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. - PubMed
-
- Groth A, Rocha W, Verreault A, Almouzni G. Chromatin challenges during DNA replication and repair. Cell. 2007;128:721–733. - PubMed
-
- Thomas JO, Travers AA. HMG1 and 2, and related ‘architectural’ DNA-binding proteins. Trends Biochem Sci. 2001;26:167–174. - PubMed
-
- Stros M. HMGB proteins: interactions with DNA and chromatin. Biochim Biophys Acta. 2010;1799:101–113. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

