Inhibition of XMRV and HIV-1 proteases by pepstatin A and acetyl-pepstatin
- PMID: 22804908
- PMCID: PMC6290463
- DOI: 10.1111/j.1742-4658.2012.08714.x
Inhibition of XMRV and HIV-1 proteases by pepstatin A and acetyl-pepstatin
Abstract
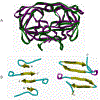
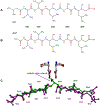
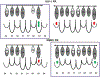
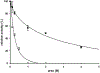
The kinetic properties of two classical inhibitors of aspartic proteases (PRs), pepstatin A and acetyl-pepstatin, were compared in their interactions with HIV-1 and xenotropic murine leukemia virus related virus (XMRV) PRs. Both compounds are substantially weaker inhibitors of XMRV PR than of HIV-1 PR. Previous kinetic and structural studies characterized HIV-1 PR-acetyl-pepstatin and XMRV PR-pepstatin A complexes and suggested dramatically different binding modes. Interaction energies were calculated for the possible binding modes and suggested a strong preference for the one-inhibitor binding mode for HIV-1 PR-acetyl-pepstatin and the two-inhibitor binding mode for XMRV PR-pepstatin A interactions. Comparison of the molecular models suggested that in the case of XMRV PR the relatively unfavorable interactions at S3' and the favorable interactions at S4 and S4' sites with the statine residues may shift the ground state binding towards the two-inhibitor binding mode, whereas the single molecule ground state binding of statines to the HIV-1 PR appear to be more favorable. The preferred single molecular binding to HIV-1 PR allows the formation of the transition state complex, represented by substantially better binding constants. Intriguingly, the crystal structure of the complex of acetyl-pepstatin with XMRV PR has shown a mixed type of binding: the unusual binding mode of two molecules of the inhibitor to the enzyme, in a mode very similar to the previously determined complex with pepstatin A, together with the classical binding mode found for HIV-1 PR. The structure is thus in good agreement with the very similar interaction energies calculated for the two types of binding.
© 2012 The Authors Journal compilation © 2012 FEBS.
Conflict of interest statement
Authors declared no conflict of interest.
Figures
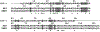

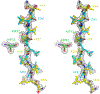
Similar articles
-
Structural and biochemical characterization of the inhibitor complexes of xenotropic murine leukemia virus-related virus protease.FEBS J. 2011 Nov;278(22):4413-24. doi: 10.1111/j.1742-4658.2011.08364.x. Epub 2011 Oct 10. FEBS J. 2011. PMID: 21951660 Free PMC article.
-
NMR study of xenotropic murine leukemia virus-related virus protease in a complex with amprenavir.Biochem Biophys Res Commun. 2012 Aug 24;425(2):284-9. doi: 10.1016/j.bbrc.2012.07.083. Epub 2012 Jul 25. Biochem Biophys Res Commun. 2012. PMID: 22842568
-
Intrinsic DNA synthesis fidelity of xenotropic murine leukemia virus-related virus reverse transcriptase.FEBS J. 2012 Apr;279(8):1433-44. doi: 10.1111/j.1742-4658.2012.08532.x. Epub 2012 Mar 16. FEBS J. 2012. PMID: 22340433 Free PMC article.
-
Pepstatin inhibition mechanism.Adv Exp Med Biol. 1977;95:199-210. doi: 10.1007/978-1-4757-0719-9_12. Adv Exp Med Biol. 1977. PMID: 339690
-
Molecular and enzymatic characterization of XMRV protease by a cell-free proteolytic analysis.J Proteomics. 2012 Aug 3;75(15):4863-73. doi: 10.1016/j.jprot.2012.05.047. Epub 2012 Jun 9. J Proteomics. 2012. PMID: 22687250
Cited by
-
Different Mutation Tolerance of Lentiviral (HIV-1) and Deltaretroviral (BLV and HTLV) Protease Precursors.Viruses. 2022 Aug 26;14(9):1888. doi: 10.3390/v14091888. Viruses. 2022. PMID: 36146695 Free PMC article.
-
Genome-Wide Identification and Expression Profiling of Glycosidases, Lipases, and Proteases from Invasive Asian Palm Weevil, Rhynchophorus ferrugineus.Insects. 2025 Apr 17;16(4):421. doi: 10.3390/insects16040421. Insects. 2025. PMID: 40332944 Free PMC article.
-
Biochemical Characterization of Human Retroviral-Like Aspartic Protease 1 (ASPRV1).Biomolecules. 2020 Jul 6;10(7):1004. doi: 10.3390/biom10071004. Biomolecules. 2020. PMID: 32640672 Free PMC article.
-
Impact of novel microbial secondary metabolites on the pharma industry.Appl Microbiol Biotechnol. 2022 Mar;106(5-6):1855-1878. doi: 10.1007/s00253-022-11821-5. Epub 2022 Feb 21. Appl Microbiol Biotechnol. 2022. PMID: 35188588 Free PMC article. Review.
-
Tandem ketone reduction in pepstatin biosynthesis reveals an F420H2-dependent statine pathway.Nat Commun. 2025 May 15;16(1):4531. doi: 10.1038/s41467-025-59785-0. Nat Commun. 2025. PMID: 40374670 Free PMC article.
References
-
- Wlodawer A & Vondrasek J (1998) Inhibitors of HIV-1 protease: A major success of structure-assisted drug design. Annu. Rev. Biophys. Biomol. Struct 27, 249–284. - PubMed
-
- Scourfield A, Waters L, Nelson M (2011) Drug combinations for HIV: what's new? Expert Rev Anti Infect Ther 9: 1001–1011. - PubMed
-
- Poiesz BJ, Ruscetti FW, Reitz MS, Kalyanaraman VS & Gallo RC (1981) Isolation of a new type C retrovirus (HTLV) in primary uncultured cells of a patient with Sezary T-cell leukaemia. Nature 294, 268–271. - PubMed
-
- Groom HC & Bishop KN (2012) The tale of xenotropic murine leukemia virus-related virus. J. Gen. Virol 93, 915–924. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

