Enhanced stability of microRNA expression facilitates classification of FFPE tumour samples exhibiting near total mRNA degradation
- PMID: 22805332
- PMCID: PMC3419950
- DOI: 10.1038/bjc.2012.294
Enhanced stability of microRNA expression facilitates classification of FFPE tumour samples exhibiting near total mRNA degradation
Abstract
Background: As degradation of formalin-fixed paraffin-embedded (FFPE) samples limits the ability to profile mRNA expression, we explored factors predicting the success of mRNA expression profiling of FFPE material and investigated an approach to overcome the limitation.
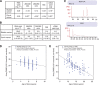
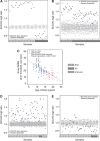
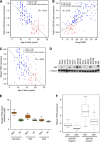
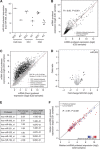
Methods: Bladder (n=140, stored 3-8 years) and cervix (n=160, stored 8-23 years) carcinoma FFPE samples were hybridised to Affymetrix Exon 1.0ST arrays. Percentage detection above background (%DABG) measured technical success. Biological signal was assessed by distinguishing cervix squamous cell carcinoma (SCC) and adenocarcinoma (AC) using a gene signature. As miR-205 had been identified as a marker of SCC, precursor mir-205 was measured by Exon array and mature miR-205 by qRT-PCR. Genome-wide microRNA (miRNA) expression (Affymetrix miRNA v2.0 arrays) was compared in eight newer FFPE samples with biological signal and eight older samples without.
Results: RNA quality controls (QCs) (e.g., RNA integrity (RIN) number) failed to predict profiling success, but sample age correlated with %DABG in bladder (R=-0.30, P<0.01) and cervix (R=-0.69, P<0.01). Biological signal was lost in older samples and neither a signature nor precursor mir-205 separated samples by histology. miR-205 qRT-PCR discriminated SCC from AC, validated by miRNA profiling (26-fold higher in SCC; P=1.10 × 10(-5)). Genome-wide miRNA (R=0.95) and small nucleolar RNA (R=0.97) expression correlated well in the eight newer vs older FFPE samples and better than mRNA expression (R=0.72).
Conclusion: Sample age is the best predictor of successful mRNA profiling of FFPE material, and miRNA profiling overcomes the limitation of age and copes well with older samples.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Abramovitz M, Barwick BG, Willis S, Young B, Catzavelos C, Li Z, Kodani M, Tang W, Bouzyk M, Moreno CS, Leyland-Jones B (2011) Molecular characterisation of formalin-fixed paraffin-embedded (FFPE) breast tumour specimens using a custom 512-gene breast cancer bead array-based platform. Br J Cancer 105: 1574–1581 - PMC - PubMed
-
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B 57: 289–300
-
- Blow N (2007) Tissue preparation: tissue issues. Nature 448: 959–963 - PubMed
-
- Cassidy SB, Schwartz S, Miller JL, Driscoll DJ (2012) Prader-Willi syndrome. Genet Med 14: 10–26 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

