Neddylation dysfunction in Alzheimer's disease
- PMID: 22805479
- PMCID: PMC3484225
- DOI: 10.1111/j.1582-4934.2012.01604.x
Neddylation dysfunction in Alzheimer's disease
Abstract
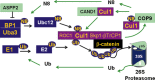
Ubiquitin-dependent proteolysis is a major mechanism that downregulates misfolded proteins or those that have finished a programmed task. In the last two decades, neddylation has emerged as a major regulatory pathway for ubiquitination. Central to the neddylation pathway is the amyloid precursor protein (APP)-binding protein APP-BP1, which together with Uba3, plays an analogous role to the ubiquitin-activating enzyme E1 in nedd8 activation. Activated nedd8 covalently modifies and activates a major class of ubiquitin ligases called Cullin-RING ligases (CRLs). New evidence suggests that neddylation also modifies Type-1 transmembrane receptors such as APP. Here we review the functions of neddylation and summarize evidence suggesting that dysfunction of neddylation is involved in Alzheimer's disease.
© 2012 The Authors Journal of Cellular and Molecular Medicine © 2012 Foundation for Cellular and Molecular Medicine/Blackwell Publishing Ltd.
Figures


References
-
- Leyser HM, Lincoln CA, Timpte C, et al. Arabidopsis auxin-resistance gene AXR1 encodes a protein related to ubiquitin-activating enzyme E1. Nature. 1993;364:161–4. - PubMed
-
- Kumar S, Yoshida Y, Noda M. Cloning of a cDNA which encodes a novel ubiquitin-like protein. Biochem Biophys Res Commun. 1993;195:393–9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

