The in planta transcriptome of Ralstonia solanacearum: conserved physiological and virulence strategies during bacterial wilt of tomato
- PMID: 22807564
- PMCID: PMC3413399
- DOI: 10.1128/mBio.00114-12
The in planta transcriptome of Ralstonia solanacearum: conserved physiological and virulence strategies during bacterial wilt of tomato
Abstract
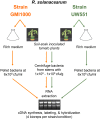
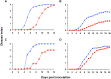
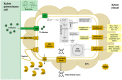
Plant xylem fluid is considered a nutrient-poor environment, but the bacterial wilt pathogen Ralstonia solanacearum is well adapted to it, growing to 10(8) to 10(9) CFU/g tomato stem. To better understand how R. solanacearum succeeds in this habitat, we analyzed the transcriptomes of two phylogenetically distinct R. solanacearum strains that both wilt tomato, strains UW551 (phylotype II) and GMI1000 (phylotype I). We profiled bacterial gene expression at ~6 × 10(8) CFU/ml in culture or in plant xylem during early tomato bacterial wilt pathogenesis. Despite phylogenetic differences, these two strains expressed their 3,477 common orthologous genes in generally similar patterns, with about 12% of their transcriptomes significantly altered in planta versus in rich medium. Several primary metabolic pathways were highly expressed during pathogenesis. These pathways included sucrose uptake and catabolism, and components of these pathways were encoded by genes in the scrABY cluster. A UW551 scrA mutant was significantly reduced in virulence on resistant and susceptible tomato as well as on potato and the epidemiologically important weed host Solanum dulcamara. Functional scrA contributed to pathogen competitive fitness during colonization of tomato xylem, which contained ~300 µM sucrose. scrA expression was induced by sucrose, but to a much greater degree by growth in planta. Unexpectedly, 45% of the genes directly regulated by HrpB, the transcriptional activator of the type 3 secretion system (T3SS), were upregulated in planta at high cell densities. This result modifies a regulatory model based on bacterial behavior in culture, where this key virulence factor is repressed at high cell densities. The active transcription of these genes in wilting plants suggests that T3SS has a biological role throughout the disease cycle. IMPORTANCE Ralstonia solanacearum is a widespread plant pathogen that causes bacterial wilt disease. It inflicts serious crop losses on tropical farmers, with major economic and human consequences. It is also a model for the many destructive microbes that colonize the water-conducting plant xylem tissue, which is low in nutrients and oxygen. We extracted bacteria from infected tomato plants and globally identified the biological functions that R. solanacearum expresses during plant pathogenesis. This revealed the unexpected presence of sucrose in tomato xylem fluid and the pathogen's dependence on host sucrose for virulence on tomato, potato, and the common weed bittersweet nightshade. Further, R. solanacearum was highly responsive to the plant environment, expressing several metabolic and virulence functions quite differently in the plant than in pure culture. These results reinforce the utility of studying pathogens in interaction with hosts and suggest that selecting for reduced sucrose levels could generate wilt-resistant crops.
Figures



Comment in
-
"Listening in" on how a bacterium takes over the plant vascular system.mBio. 2012 Sep 11;3(5):e00269-12. doi: 10.1128/mBio.00269-12. Print 2012. mBio. 2012. PMID: 22967979 Free PMC article.
References
-
- Pegg GF. 1985. Life in a black hole: the microenvironment of the vascular pathogen. Trans. Br. Mycol. Soc. 85:1–20
-
- Denny TP. 2006. Plant pathogenic Ralstonia species, p 573–644 In Plant-associated bacteria. Springer Verlag, Dordrecht, The Netherlands
-
- Prior P, Fegan M. 2005. Recent developments in the phylogeny and classification of Ralstonia solanacearum. Acta Hort. 695:127–136
-
- Gabriel DW, et al. 2006. Identification of open reading frames unique to a select agent: Ralstonia solanacearum race 3 biovar 2. Mol. Plant Microbe Interact. 19:69–79 - PubMed
-
- Milling A, Meng F, Denny TP, Allen C. 2009. Interactions with hosts at cool temperatures, not cold tolerance, explain the unique epidemiology of Ralstonia solanacearum race 3 biovar 2. Phytopathology 99:1127–1134 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
