Transposon-derived and satellite-derived repetitive sequences play distinct functional roles in Mammalian intron size expansion
- PMID: 22807622
- PMCID: PMC3396637
- DOI: 10.4137/EBO.S9758
Transposon-derived and satellite-derived repetitive sequences play distinct functional roles in Mammalian intron size expansion
Abstract
Background: Repetitive sequences (RSs) are redundant, complex at times, and often lineage-specific, representing significant "building" materials for genes and genomes. According to their origins, sequence characteristics, and ways of propagation, repetitive sequences are divided into transposable elements (TEs) and satellite sequences (SSs) as well as related subfamilies and subgroups hierarchically. The combined changes attributable to the repetitive sequences alter gene and genome architectures, such as the expansion of exonic, intronic, and intergenic sequences, and most of them propagate in a seemingly random fashion and contribute very significantly to the entire mutation spectrum of mammalian genomes.
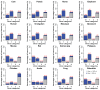
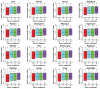
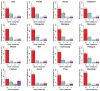
Principal findings: Our analysis is focused on evolutional features of TEs and SSs in the intronic sequence of twelve selected mammalian genomes. We divided them into four groups-primates, large mammals, rodents, and primary mammals-and used four non-mammalian vertebrate species as the out-group. After classifying intron size variation in an intron-centric way based on RS-dominance (TE-dominant or SS-dominant intron expansions), we observed several distinct profiles in intron length and positioning in different vertebrate lineages, such as retrotransposon-dominance in mammals and DNA transposon-dominance in the lower vertebrates, amphibians and fishes. The RS patterns of mouse and rat genes are most striking, which are not only distinct from those of other mammals but also different from that of the third rodent species analyzed in this study-guinea pig. Looking into the biological functions of relevant genes, we observed a two-dimensional divergence; in particular, genes that possess SS-dominant and/or RS-free introns are enriched in tissue-specific development and transcription regulation in all mammalian lineages. In addition, we found that the tendency of transposons in increasing intron size is much stronger than that of satellites, and the combined effect of both RSs is greater than either one of them alone in a simple arithmetic sum among the mammals and the opposite is found among the four non-mammalian vertebrates.
Conclusions: TE- and SS-derived RSs represent major mutational forces shaping the size and composition of vertebrate genes and genomes, and through natural selection they either fine-tune or facilitate changes in size expansion, position variation, and duplication, and thus in functions and evolutionary paths for better survival and fitness. When analyzed globally, not only are such changes significantly diversified but also comprehensible in lineages and biological implications.
Keywords: intron size; mammalian genomes; satellite sequences; transposable elements.
Figures





References
LinkOut - more resources
Full Text Sources

